Functional interaction between the coactivator Drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers
- PMID: 11094082
- PMCID: PMC102188
- DOI: 10.1128/MCB.20.24.9317-9330.2000
Functional interaction between the coactivator Drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers
Abstract
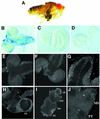
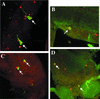
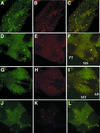
CREB-binding protein (CBP) is a coactivator for multiple transcription factors that transduce a variety of signaling pathways. Current models propose that CBP enhances gene expression by bridging the signal-responsive transcription factors with components of the basal transcriptional machinery and by augmenting the access of transcription factors to DNA through the acetylation of histones. To define the pathways and proteins that require CBP function in a living organism, we have begun a genetic analysis of CBP in flies. We have overproduced Drosophila melanogaster CBP (dCBP) in a variety of cell types and obtained distinct adult phenotypes. We used an uninflated-wing phenotype, caused by the overexpression of dCBP in specific central nervous system cells, to screen for suppressors of dCBP overactivity. Two genes with mutant versions that act as dominant suppressors of the wing phenotype were identified: the PKA-C1/DCO gene, encoding the catalytic subunit of cyclic AMP protein kinase, and ash1, a member of the trithorax group (trxG) of chromatin modifiers. Using immunocolocalization, we showed that the ASH1 protein is specifically expressed in the majority of the dCBP-overexpressing cells, suggesting that these proteins have the potential to interact biochemically. This model was confirmed by the findings that the proteins interact strongly in vitro and colocalize at specific sites on polytene chromosomes. The trxG proteins are thought to maintain gene expression during development by creating domains of open chromatin structure. Our results thus implicate a second class of chromatin-associated proteins in mediating dCBP function and imply that dCBP might be involved in the regulation of higher-order chromatin structure.
Figures
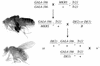




References
-
- Akimaru H, Chen Y, Dai P, Hou D-X, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386:735–738. - PubMed
-
- Akimaru H, Hou D-X, Ishii S. Drosophila CBP is required for dorsal-dependent twist gene expression. Nat Genet. 1998;17:211–214. - PubMed
-
- Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. - PubMed
-
- Aza-Blanc P, Ramirez-Weber F-A, Laget M-P, Schwartz C, Kornberg T. Proteolysis that is inhibited by Hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
