Cisplatin-DNA adducts inhibit translocation of the Ku subunits of DNA-PK
- PMID: 11095672
- PMCID: PMC115169
- DOI: 10.1093/nar/28.23.4634
Cisplatin-DNA adducts inhibit translocation of the Ku subunits of DNA-PK
Abstract
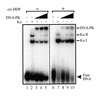
We have determined the effect of cisplatin-DNA damage on the ability of the DNA-dependent protein kinase (DNA-PK) to interact with duplex DNA molecules in vitro. The Ku DNA binding subunits of DNA-PK display a reduced ability to translocate on duplex DNA containing cisplatin-DNA adducts compared to control, undamaged duplex DNA. The decreased rates of translocation resulted in a decrease in the association of the p460 catalytic subunit of DNA-PK (DNA-PKcs) with the Ku-DNA complex. In addition to a decrease in DNA-PKcs association, the DNA-PKcs that is bound with Ku at a DNA end containing cisplatin-DNA adducts has a reduced catalytic rate compared to heterotrimeric DNA-PK assembled on undamaged DNA. The position of the cisplatin-DNA lesion from the terminus also effects kinase activation, with maximal inhibition occurring when the lesion is closer to the terminus. These results are consistent with a model for DNA-PK activation where the Ku dimer translocates away from the DNA terminus and facilitates the association of DNA-PKcs which interacts with both Ku and DNA resulting in kinase activation. The presence of cisplatin adducts decreases the ability to translocate away from the terminus and results in the formation of inactive kinase complexes at the DNA terminus. The results are discussed with respect to the ability of cisplatin to sensitize cells to DNA damage induced by ionizing radiation and the ability to repair DNA double-strand breaks.
Figures






Similar articles
-
Human Ku autoantigen binds cisplatin-damaged DNA but fails to stimulate human DNA-activated protein kinase.J Biol Chem. 1996 Jun 7;271(23):13861-7. doi: 10.1074/jbc.271.23.13861. J Biol Chem. 1996. PMID: 8662830
-
Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein.Nucleic Acids Res. 1999 Dec 15;27(24):4679-86. doi: 10.1093/nar/27.24.4679. Nucleic Acids Res. 1999. PMID: 10572166 Free PMC article.
-
Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment.J Mol Biol. 2003 Feb 7;326(1):93-103. doi: 10.1016/s0022-2836(02)01328-1. J Mol Biol. 2003. PMID: 12547193
-
Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids.Nucleic Acids Res. 1998 Apr 1;26(7):1551-9. doi: 10.1093/nar/26.7.1551. Nucleic Acids Res. 1998. PMID: 9512523 Free PMC article. Review.
-
Regulation and repair of double-strand DNA breaks.Crit Rev Eukaryot Gene Expr. 1996;6(4):345-75. doi: 10.1615/critreveukargeneexpr.v6.i4.20. Crit Rev Eukaryot Gene Expr. 1996. PMID: 8959372 Review.
Cited by
-
"An End to a Means": How DNA-End Structure Shapes the Double-Strand Break Repair Process.Front Mol Biosci. 2020 Jan 10;6:153. doi: 10.3389/fmolb.2019.00153. eCollection 2019. Front Mol Biosci. 2020. PMID: 31998749 Free PMC article. Review.
-
DNA-dependent conformational changes in the Ku heterodimer.Biochemistry. 2008 Apr 15;47(15):4359-68. doi: 10.1021/bi702284c. Epub 2008 Mar 21. Biochemistry. 2008. PMID: 18355052 Free PMC article.
-
Suppression of nonhomologous end joining repair by overexpression of HMGA2.Cancer Res. 2009 Jul 15;69(14):5699-706. doi: 10.1158/0008-5472.CAN-08-4833. Epub 2009 Jun 23. Cancer Res. 2009. PMID: 19549901 Free PMC article.
-
Coordination of DNA-PK activation and nuclease processing of DNA termini in NHEJ.Antioxid Redox Signal. 2011 Jun 15;14(12):2531-43. doi: 10.1089/ars.2010.3368. Epub 2010 Dec 2. Antioxid Redox Signal. 2011. PMID: 20698792 Free PMC article. Review.
-
Cisplatin Reduces the Frequencies of Radiotherapy-Induced Micronuclei in Peripheral Blood Lymphocytes of Patients with Gynaecological Cancer: Possible Implications for the Risk of Second Malignant Neoplasms.Cells. 2021 Oct 9;10(10):2709. doi: 10.3390/cells10102709. Cells. 2021. PMID: 34685687 Free PMC article.
References
-
- Yoo S., Kimzey,A. and Dynan,W.S. (1999) Photocross-Linking of an Oriented DNA Repair Complex — Ku Bound at a Single DNA End. J. Biol. Chem., 274, 20034–20039. - PubMed
-
- de Vries E.G., van Driel,W., Bergsma,W.G., Arnberg,A.C. and van der Vliet,P.C. (1989) HeLa Nuclear Protein Recognizing DNA Termini and Translocating on DNA Forming a Regular DNA-Multimeric Protein Complex. J. Mol. Biol., 208, 65–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

