CIRP2, a major cytoplasmic RNA-binding protein in Xenopus oocytes
- PMID: 11095679
- PMCID: PMC115157
- DOI: 10.1093/nar/28.23.4689
CIRP2, a major cytoplasmic RNA-binding protein in Xenopus oocytes
Abstract
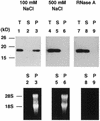
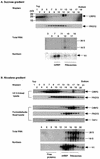
In an attempt to isolate mRNA-binding proteins we fractionated Xenopus oocyte lysate by oligo(dT)-cellulose chromatography. A 20 kDa protein was the major component of the eluate. cDNA cloning revealed that this protein is a Xenopus homolog of the cold-inducible RNA-binding protein (CIRP) which was originally identified in mammalian cells as a protein that is overexpressed upon a temperature downshift. This Xenopus protein, termed here xCIRP2, is highly expressed in ovary, testis and brain in adult Xenopus tissues. In oocytes it is predominantly localized in the cytoplasm. By biochemical fractionation we provide evidence that xCIRP2 is associated with ribosomes, suggesting that it participates in translational regulation in oocytes. Microinjection of labeled mRNA into oocytes followed by UV cross-linking of the oocyte lysate led to identification of two major RNA-binding activities. Immunoprecipitation of the RNA-binding proteins demonstrated that one is xCIRP2 and that the other contains FRGY2. FRGY2, which is one of the principal constituents of mRNA storage particles involved in translational masking of maternal mRNA, has an RNA-binding domain conserved to those of bacterial cold shock proteins. Possible implications of the highly abundant expression in oocytes of cold shock RNA-binding proteins of both eukaryotic and prokaryotic types are discussed.
Figures





Similar articles
-
Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution.Nucleic Acids Res. 2002 Dec 1;30(23):5182-92. doi: 10.1093/nar/gkf638. Nucleic Acids Res. 2002. PMID: 12466543 Free PMC article.
-
Xenopus cold-inducible RNA-binding protein 2 interacts with ElrA, the Xenopus homolog of HuR, and inhibits deadenylation of specific mRNAs.J Biol Chem. 2003 Nov 28;278(48):48491-7. doi: 10.1074/jbc.M308328200. Epub 2003 Sep 17. J Biol Chem. 2003. PMID: 13679363
-
Identification of four CCCH zinc finger proteins in Xenopus, including a novel vertebrate protein with four zinc fingers and severely restricted expression.Gene. 1999 Mar 4;228(1-2):133-45. doi: 10.1016/s0378-1119(98)00617-9. Gene. 1999. PMID: 10072766
-
A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking.Scand J Clin Lab Invest Suppl. 2001;234:93-9. Scand J Clin Lab Invest Suppl. 2001. PMID: 11713986 Review.
-
Coupling transcription to translation: a novel site for the regulation of eukaryotic gene expression.Int J Biochem Cell Biol. 1996 Mar;28(3):247-57. doi: 10.1016/1357-2725(95)00141-7. Int J Biochem Cell Biol. 1996. PMID: 8920634 Review.
Cited by
-
Molecular phenotype of zebrafish ovarian follicle by serial analysis of gene expression and proteomic profiling, and comparison with the transcriptomes of other animals.BMC Genomics. 2006 Mar 9;7:46. doi: 10.1186/1471-2164-7-46. BMC Genomics. 2006. PMID: 16526958 Free PMC article.
-
Cold-inducible RNA-binding protein migrates from the nucleus to the cytoplasm under cold stress in normal human bronchial epithelial cells via TRPM8-mediated mechanism.Ann Transl Med. 2021 Sep;9(18):1470. doi: 10.21037/atm-21-4447. Ann Transl Med. 2021. PMID: 34734022 Free PMC article.
-
Compaction of RNA Duplexes in the Cell*.Angew Chem Int Ed Engl. 2020 Dec 14;59(51):23025-23029. doi: 10.1002/anie.202009800. Epub 2020 Oct 13. Angew Chem Int Ed Engl. 2020. PMID: 32804430 Free PMC article.
-
Cross-species hybridizations on a multi-species cDNA microarray to identify evolutionarily conserved genes expressed in oocytes.BMC Genomics. 2006 May 10;7:113. doi: 10.1186/1471-2164-7-113. BMC Genomics. 2006. PMID: 16686947 Free PMC article.
-
Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis.Proc Natl Acad Sci U S A. 2005 Feb 8;102(6):1865-70. doi: 10.1073/pnas.0409764102. Epub 2005 Jan 31. Proc Natl Acad Sci U S A. 2005. PMID: 15684048 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

