Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA
- PMID: 11095680
- PMCID: PMC115172
- DOI: 10.1093/nar/28.23.4698
Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA
Abstract
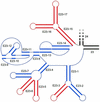
The secondary structure of V4, the largest variable area of eukaryotic small subunit ribosomal RNA, was re-examined by comparative analysis of 3253 nucleotide sequences distributed over the animal, plant and fungal kingdoms and a diverse set of protist taxa. An extensive search for compensating base pair substitutions and for base covariation revealed that in most eukaryotes the secondary structure of the area consists of 11 helices and includes two pseudoknots. In one of the pseudoknots, exchange of base pairs between the two stems seems to occur, and covariation analysis points to the presence of a base triple. The area also contains three potential insertion points where additional hairpins or branched structures are present in a number of taxa scattered throughout the eukaryotic domain.
Figures





References
-
- Gutell R.R., Weiser,B., Woese,C.R. and Noller,H.F. (1985) Progr. Nucleic Acids Res. Mol. Biol., 32, 155–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

