Fission yeast retrotransposon Tf1 integration is targeted to 5' ends of open reading frames
- PMID: 11095681
- PMCID: PMC115174
- DOI: 10.1093/nar/28.23.4709
Fission yeast retrotransposon Tf1 integration is targeted to 5' ends of open reading frames
Abstract
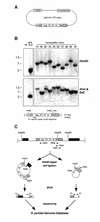
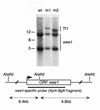
Target site selection of transposable elements is usually not random but involves some specificity for a DNA sequence or a DNA binding host factor. We have investigated the target site selection of the long terminal repeat-containing retrotransposon Tf1 from the fission yeast Schizosaccharomyces pombe. By monitoring induced transposition events we found that Tf1 integration sites were distributed throughout the genome. Mapping these insertions revealed that Tf1 did not integrate into open reading frames, but occurred preferentially in longer intergenic regions with integration biased towards a region 100-420 bp upstream of the translation start site. Northern blot analysis showed that transcription of genes adjacent to Tf1 insertions was not significantly changed.
Figures


References
-
- Craig N.L. (1997) Annu. Rev. Biochem., 66, 437–474. - PubMed
-
- Engels W.R. (1996) Curr. Top. Microbiol. Immunol., 204, 103–123. - PubMed
-
- Kim J.M., Vanguri,S., Boeke,J.D., Gabriel,A. and Voytas,D.F. (1998) Genome Res., 8, 464–478. - PubMed
-
- Bryk M., Banerjee,M., Murphy,M., Knudsen,K.E., Garfinkel,D.J. and Curcio,M.J. (1997) Genes Dev., 11, 255–269. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

