Error-prone lesion bypass by human DNA polymerase eta
- PMID: 11095682
- PMCID: PMC115171
- DOI: 10.1093/nar/28.23.4717
Error-prone lesion bypass by human DNA polymerase eta
Abstract
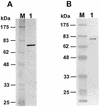
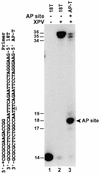
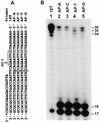
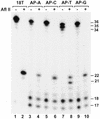
DNA lesion bypass is an important cellular response to genomic damage during replication. Human DNA polymerase eta (Pol(eta)), encoded by the Xeroderma pigmentosum variant (XPV) gene, is known for its activity of error-free translesion synthesis opposite a TT cis-syn cyclobutane dimer. Using purified human Pol(eta), we have examined bypass activities of this polymerase opposite several other DNA lesions. Human Pol(eta) efficiently bypassed a template 8-oxoguanine, incorporating an A or a C opposite the lesion with similar efficiencies. Human Pol(eta) effectively bypassed a template abasic site, incorporating an A and less frequently a G opposite the lesion. Significant -1 deletion was also observed when the template base 5' to the abasic site is a T. Human Pol(eta) partially bypassed a template (+)-trans-anti-benzo[a]pyrene-N:(2)-dG and predominantly incorporated an A, less frequently a T, and least frequently a G or a C opposite the lesion. This specificity of nucleotide incorporation correlates well with the known mutation spectrum of (+)-trans-anti-benzo[a]pyrene-N:(2)-dG lesion in mammalian cells. These results show that human Pol(eta) is capable of error-prone translesion DNA syntheses in vitro and suggest that Pol(eta) may bypass certain lesions with a mutagenic consequence in humans.
Figures



References
-
- Lemontt J.F. (1977) Pathways of ultraviolet mutability in Saccharomyces cerevisiae. III. Genetic analysis and properties of mutants resistant to ultraviolet-induced forward mutation. Mutat. Res., 43, 179–204. - PubMed
-
- Lawrence C. (1994) The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays, 16, 253–258. - PubMed
-
- Lawrence C.W. and Hinkle,D.C. (1996) DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv., 28, 21–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

