The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase
- PMID: 11095689
- PMCID: PMC115166
- DOI: 10.1093/nar/28.23.4769
The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase
Abstract
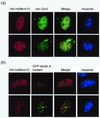
Mcm10 (Dna43), first identified in Saccharomyces cerevisiae, is an essential protein which functions in the initiation of DNA synthesis. Mcm10 is a nuclear protein that is localized to replication origins and mediates the interaction of the Mcm2-7 complex with replication origins. We identified and cloned a human cDNA whose product was structurally homologous to the yeast Mcm10 protein. Human Mcm10 (HsMcm10) is a 98-kDa protein of 874 amino acids which shows 23 and 21% overall similarity to Schizosaccharomyces pombe Cdc23 and S. cerevisiae Mcm10, respectively. The messenger RNA level of HsMcm10 increased at the G(1)/S-boundary when quiescent human NB1-RGB cells were induced to proliferate as is the case of many replication factors. HsMcm10 associated with nuclease-resistant nuclear structures throughout S phase and dissociated from it in G(2) phase. HsMcm10 associated with human Orc2 protein when overexpressed in COS-1 cells. HsMcm10 also interacted with Orc2, Mcm2 and Mcm6 proteins in the yeast two-hybrid system. These results suggest that HsMcm10 may function in DNA replication through the interaction with Orc and Mcm2-7 complexes.
Figures





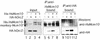
Similar articles
-
The essential schizosaccharomyces pombe cdc23 DNA replication gene shares structural and functional homology with the Saccharomyces cerevisiae DNA43 (MCM10) gene.Curr Genet. 1998 Sep;34(3):164-71. doi: 10.1007/s002940050382. Curr Genet. 1998. PMID: 9745018
-
Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins.Genes Dev. 2000 Apr 15;14(8):913-26. Genes Dev. 2000. PMID: 10783164 Free PMC article.
-
Fission yeast Cdc23 interactions with DNA replication initiation proteins.Curr Genet. 2002 Aug;41(5):342-8. doi: 10.1007/s00294-002-0316-9. Epub 2002 Jul 23. Curr Genet. 2002. PMID: 12185500
-
A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae.Mol Cell Biol. 1997 Jun;17(6):3261-71. doi: 10.1128/MCB.17.6.3261. Mol Cell Biol. 1997. PMID: 9154825 Free PMC article.
-
Initiation of DNA replication in eukaryotic cells.Annu Rev Cell Dev Biol. 1997;13:293-332. doi: 10.1146/annurev.cellbio.13.1.293. Annu Rev Cell Dev Biol. 1997. PMID: 9442876 Review.
Cited by
-
Human SIRT1 regulates DNA binding and stability of the Mcm10 DNA replication factor via deacetylation.Nucleic Acids Res. 2013 Apr;41(7):4065-79. doi: 10.1093/nar/gkt131. Epub 2013 Feb 28. Nucleic Acids Res. 2013. PMID: 23449222 Free PMC article.
-
Ten-Eleven Translocation 1 and 2 Confer Overlapping Transcriptional Programs for the Proliferation of Cultured Adult Neural Stem Cells.Cell Mol Neurobiol. 2017 Aug;37(6):995-1008. doi: 10.1007/s10571-016-0432-6. Epub 2016 Oct 24. Cell Mol Neurobiol. 2017. PMID: 27778125 Free PMC article.
-
Structural basis for DNA binding by replication initiator Mcm10.Structure. 2008 Dec 10;16(12):1892-901. doi: 10.1016/j.str.2008.10.005. Structure. 2008. PMID: 19081065 Free PMC article.
-
Structural biology of replication initiation factor Mcm10.Subcell Biochem. 2012;62:197-216. doi: 10.1007/978-94-007-4572-8_11. Subcell Biochem. 2012. PMID: 22918587 Free PMC article. Review.
-
The CENP-B homolog, Abp1, interacts with the initiation protein Cdc23 (MCM10) and is required for efficient DNA replication in fission yeast.Cell Div. 2006 Nov 17;1:27. doi: 10.1186/1747-1028-1-27. Cell Div. 2006. PMID: 17112379 Free PMC article.
References
-
- Bell S.P. and Stillman,B. (1992) Nature, 357, 128–134. - PubMed
-
- Diffley J.F.X. and Cocker,J.H. (1992) Nature, 357, 169–172. - PubMed
-
- Coleman T.R., Carpenter,P.B. and Dunphy,W.G. (1996) Cell, 87, 53–63. - PubMed
-
- Romanowski P., Madine,M.A., Rowles,A., Blow,J.J. and Laskey,R.A. (1996) Curr. Biol., 6, 1416–1425. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

