A continuous assay for DNA cleavage: the application of "break lights" to enediynes, iron-dependent agents, and nucleases
- PMID: 11095715
- PMCID: PMC17611
- DOI: 10.1073/pnas.240460997
A continuous assay for DNA cleavage: the application of "break lights" to enediynes, iron-dependent agents, and nucleases
Abstract
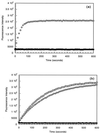
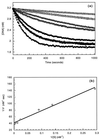
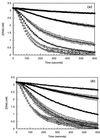
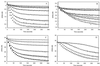
Although extensive effort has been applied toward understanding the mechanism by which enediynes cleave DNA, a continuous assay for this phenomenon is still lacking. In fact, with the exception of assays for DNase, continuous assays for most DNA cleavage events are unavailable. This article describes the application of "molecular break lights" (a single-stranded oligonucleotide that adopts a stem-and-loop structure and carries a 5'-fluorescent moiety, a 3'-nonfluorescent quenching moiety, and an appropriate cleavage site within the stem) to develop the first continuous assay for cleavage of DNA by enediynes. Furthermore, the generality of this approach is demonstrated by using the described assay to directly compare the DNA cleavage by naturally occurring enediynes [calicheamicin and esperamicin), non-enediyne small molecule agents (bleomycin, methidiumpropyl-EDTA-Fe(II), and EDTA-Fe(II]), as well as the restriction endonuclease BamHI. Given the simplicity, speed, and sensitivity of this approach, the described methodology could easily be extended to a high throughput format and become a new method of choice in modern drug discovery to screen for novel protein-based or small molecule-derived DNA cleavage agents.
Figures


References
-
- Thorson, J. S., Sievers, E. L., Ahlert, J., Shepard, E., Onwueme, K. C. & Ruppen, M. (2000) Curr. Pharm. Des., in press. - PubMed
-
- Thorson J S, Shen B, Whitwam R E, Liu W, Li Y, Ahlert J. Bioorgan Chem. 1999;27:172–188.
-
- Borders D B, Doyle T W. Enediyne Antibiotics as Antitumor Agents. New York: Dekker; 1995.
-
- Smith A L, Nicolaou K C. J Med Chem. 1996;39:2103–2117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

