Coevolution of transcriptional and allosteric regulation at the chorismate metabolic branch point of Saccharomyces cerevisiae
- PMID: 11095720
- PMCID: PMC17619
- DOI: 10.1073/pnas.240469697
Coevolution of transcriptional and allosteric regulation at the chorismate metabolic branch point of Saccharomyces cerevisiae
Abstract
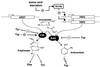
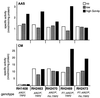
Control of transcription and enzyme activities are two interwoven regulatory systems essential for the function of a metabolic node. Saccharomyces cerevisiae strains differing in enzyme activities at the chorismate branch point of aromatic amino acid biosynthesis were constructed by recombinant DNA technology. Expression of an allosterically unregulated, constitutively activated chorismate mutase encoded by the ARO7(T226I) (ARO7(c)) allele depleted the chorismate pool. The resulting tryptophan limitation caused growth defects, which could be counteracted only by transcriptional induction of TRP2 encoding the competing enzyme anthranilate synthase. ARO7 expression is not transcriptionally regulated by amino acids. Transcriptional activation of the ARO7(c) allele led to stronger growth retardation upon tryptophan limitation. The same effect was achieved by removing the competing enzyme anthranilate synthase, which is encoded by the TRP2 gene, from the transcriptional control. The allelic situation of ARO7(c) being under general control instead of TRP2 resulted in severe growth defects when cells were starved for tryptophan. In conclusion, the specific regulatory pattern acting on enzymatic activities at the first metabolic node of aromatic amino acid biosynthesis is necessary to maintain proper flux distribution. Therefore, the evolution of the sophisticated allosteric regulation of yeast chorismate mutase requires as prerequisite (i) that the encoding ARO7 gene is not transcriptionally regulated, whereas (ii) the transcription of the competing feedback-regulated anthranilate synthase-encoding gene is controlled by availability of amino acids.
Figures

Similar articles
-
A single point mutation results in a constitutively activated and feedback-resistant chorismate mutase of Saccharomyces cerevisiae.J Bacteriol. 1989 Mar;171(3):1245-53. doi: 10.1128/jb.171.3.1245-1253.1989. J Bacteriol. 1989. PMID: 2646272 Free PMC article.
-
A GCN4 protein recognition element is not sufficient for GCN4-dependent regulation of transcription in the ARO7 promoter of Saccharomyces cerevisiae.Mol Gen Genet. 1990 Oct;224(1):57-64. doi: 10.1007/BF00259451. Mol Gen Genet. 1990. PMID: 2277632
-
Regulation of chorismate mutase in Saccharomyces cerevisiae.Mol Gen Genet. 1990 Jan;220(2):283-8. doi: 10.1007/BF00260495. Mol Gen Genet. 1990. PMID: 2183005
-
Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway.Microbiol Rev. 1991 Sep;55(3):349-70. doi: 10.1128/mr.55.3.349-370.1991. Microbiol Rev. 1991. PMID: 1943992 Free PMC article. Review.
-
[Genetic regulatory mechanisms of amino acid biosynthesis in Saccharomyces cerevisiae: mechanism of translational control of GCN4].Tanpakushitsu Kakusan Koso. 1994 Mar;39(4):530-41. Tanpakushitsu Kakusan Koso. 1994. PMID: 8165298 Review. Japanese. No abstract available.
Cited by
-
The diversity of allosteric controls at the gateway to aromatic amino acid biosynthesis.Protein Sci. 2013 Apr;22(4):395-404. doi: 10.1002/pro.2233. Epub 2013 Mar 8. Protein Sci. 2013. PMID: 23400945 Free PMC article. Review.
-
Evolution of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase-encoding genes in the yeast Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2005 Jul 12;102(28):9784-9. doi: 10.1073/pnas.0504238102. Epub 2005 Jun 29. Proc Natl Acad Sci U S A. 2005. PMID: 15987779 Free PMC article.
-
Refined molecular hinge between allosteric and catalytic domain determines allosteric regulation and stability of fungal chorismate mutase.Proc Natl Acad Sci U S A. 2002 May 14;99(10):6631-6. doi: 10.1073/pnas.092130899. Epub 2002 May 7. Proc Natl Acad Sci U S A. 2002. PMID: 11997452 Free PMC article.
-
Metabolic engineering of Saccharomyces cerevisiae for hydroxytyrosol overproduction directly from glucose.Microb Biotechnol. 2022 May;15(5):1499-1510. doi: 10.1111/1751-7915.13957. Epub 2021 Oct 24. Microb Biotechnol. 2022. PMID: 34689412 Free PMC article.
-
Yarrowia lipolytica chassis strains engineered to produce aromatic amino acids via the shikimate pathway.Microb Biotechnol. 2021 Nov;14(6):2420-2434. doi: 10.1111/1751-7915.13745. Epub 2020 Dec 30. Microb Biotechnol. 2021. PMID: 33438818 Free PMC article.
References
-
- Hinnebusch A. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 319–414.
-
- Romero R M, Roberts M F, Phillipson J D. Phytochemistry. 1995;40:1015–1025. - PubMed
-
- Andrews P R, Smith G D, Young I G. Biochemistry. 1973;12:3492–3498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

