Lactase haplotype diversity in the Old World
- PMID: 11095994
- PMCID: PMC1234910
- DOI: 10.1086/316924
Lactase haplotype diversity in the Old World
Abstract
Lactase persistence, the genetic trait in which intestinal lactase activity persists at childhood levels into adulthood, varies in frequency in different human populations, being most frequent in northern Europeans and certain African and Arabian nomadic tribes, who have a history of drinking fresh milk. Selection is likely to have played an important role in establishing these different frequencies since the development of agricultural pastoralism approximately 9,000 years ago. We have previously shown that the element responsible for the lactase persistence/nonpersistence polymorphism in humans is cis-acting to the lactase gene and that lactase persistence is associated, in Europeans, with the most common 70-kb lactase haplotype, A. We report here a study of the 11-site haplotype in 1,338 chromosomes from 11 populations that differ in lactase persistence frequency. Our data show that haplotype diversity was generated both by point mutations and recombinations. The four globally common haplotypes (A, B, C, and U) are not closely related and have different distributions; the A haplotype is at high frequencies only in northern Europeans, where lactase persistence is common; and the U haplotype is virtually absent from Indo-European populations. Much more diversity is seen in sub-Saharan Africans than in non-Africans, consistent with an "Out of Africa" model for peopling of the Old World. Analysis of recent recombinant haplotypes by allele-specific PCR, along with deduction of the root haplotype from chimpanzee sequence, allowed construction of a haplotype network that assisted in evaluation of the relative roles of drift and selection in establishing the haplotype frequencies in the different populations. We suggest that genetic drift was important in shaping the general pattern of non-African haplotype diversity, with recent directional selection in northern Europeans for the haplotype associated with lactase persistence.
Figures
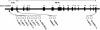
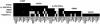



References
Electronic-Database Information
-
- Authors' “Lactase” page, http://www.gene.ucl.ac.uk/mucin/lactase.html (for AS-PCR primers)
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for 1 kb of contiguous chimpanzee sequence 5′ to the lactase gene)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for major histocompatability complex [MIM 142857, MIM 604305] and as-yet-unidentified cis-acting element [MIM 223100])
-
- UK-MRC HGMP, http://www.hgmp.mrc.ac.uk/Registered/Option/eh.html (for EH and ASSOCIATE programs)
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

