AT-rich palindromes mediate the constitutional t(11;22) translocation
- PMID: 11095996
- PMCID: PMC1234939
- DOI: 10.1086/316952
AT-rich palindromes mediate the constitutional t(11;22) translocation
Abstract
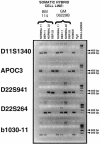
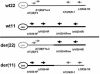
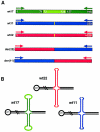
The constitutional t(11;22) translocation is the only known recurrent non-Robertsonian translocation in humans. Offspring are susceptible to der(22) syndrome, a severe congenital anomaly disorder caused by 3&rcolon;1 meiotic nondisjunction events. We previously localized the t(11;22) translocation breakpoint to a region on 22q11 within a low-copy repeat termed "LCR22" and within an AT-rich repeat on 11q23. The LCR22s are implicated in mediating different rearrangements on 22q11, leading to velocardiofacial syndrome/DiGeorge syndrome and cat-eye syndrome by homologous recombination mechanisms. The LCR22s contain AT-rich repetitive sequences, suggesting that such repeats may mediate the t(11;22) translocation. To determine the molecular basis of the translocation, we cloned and sequenced the t(11;22) breakpoint in the derivative 11 and 22 chromosomes in 13 unrelated carriers, including two de novo cases and der(22) syndrome offspring. We found that, in all cases examined, the reciprocal exchange occurred between similar AT-rich repeats on both chromosomes 11q23 and 22q11. To understand the mechanism, we examined the sequence of the breakpoint intervals in the derivative chromosomes and compared this with the deduced normal chromosomal sequence. A palindromic AT-rich sequence with a near-perfect hairpin could form, by intrastrand base-pairing, on the parental chromosomes. The sequence of the breakpoint junction in both derivatives indicates that the exchange events occurred at the center of symmetry of the palindromes, and this resulted in small, overlapping staggered deletions in this region among the different carriers. On the basis of previous studies performed in diverse organisms, we hypothesize that double-strand breaks may occur in the center of the palindrome, the tip of the putative hairpin, leading to illegitimate recombination events between similar AT-rich sequences on chromosomes 11 and 22, resulting in deletions and loss of the palindrome, which then could stabilize the DNA structure.
Figures



References
Electronic-Database Information
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for available genomic sequence on chromosomes 11q23 [AC007707] and 17 [AC004526])
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for VCFS/DGS [MIM 192430/MIM 188400] and CES [MIM 115470])
References
-
- Baumer A, Dutly F, Balmer D, Riegel M, Tukel T, Krajewska-Walasek M, Schinzel AA (1998) High level of unequal meiotic crossovers at the origin of the 22q11. 2 and 7q11.23 deletions. Hum Mol Genet 7:887–894 - PubMed
-
- Budarf ML, Collins J, Gong W, Roe B, Wang Z, Bailey LC, Sellinger B, Michaud D, Driscoll DA, Emanuel BS (1995) Cloning a balanced translocation associated with DiGeorge syndrome and identification of a disrupted candidate gene. Nat Genet 10:269–278 - PubMed
-
- Calabretta B, Robberson DL, Barrera-Saldana HA, Lambrou TP, Saunders GF (1982) Genome instability in a region of human DNA enriched in Alu repeat sequences. Nature 296:219–225 - PubMed
-
- Campana M, Serra A, Neri G (1986) Role of chromosome aberrations in recurrent abortion: a study of 269 balanced translocations. Am J Med Genet 24:341–356 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

