Role of histone acetylation in the assembly and modulation of chromatin structures
- PMID: 11097424
- PMCID: PMC5964959
- DOI: 10.3727/000000001783992687
Role of histone acetylation in the assembly and modulation of chromatin structures
Abstract
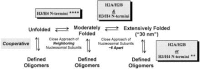
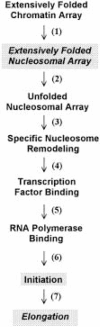
The acetylation of the core histone N-terminal "tail" domains is now recognized as a highly conserved mechanism for regulating chromatin functional states. The following article examines possible roles of acetylation in two critically important cellular processes: replication-coupled nucleosome assembly, and reversible transitions in chromatin higher order structure. After a description of the acetylation of newly synthesized histones, and of the likely acetyltransferases involved, an overview of histone octamer assembly is presented. Our current understanding of the factors thought to assemble chromatin in vivo is then described. Genetic and biochemical investigations of the function the histone tails, and their acetylation, in nucleosome assembly are detailed, followed by an analysis of the importance of histone deacetylation in the maturation of newly replicated chromatin. In the final section the involvement of the histone tail domains in chromatin higher order structures is addressed, along with the role of histone acetylation in chromatin folding. Suggestions for future research are offered in the concluding remarks.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
