Cofactor regeneration by a soluble pyridine nucleotide transhydrogenase for biological production of hydromorphone
- PMID: 11097884
- PMCID: PMC92438
- DOI: 10.1128/AEM.66.12.5161-5166.2000
Cofactor regeneration by a soluble pyridine nucleotide transhydrogenase for biological production of hydromorphone
Abstract
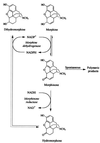
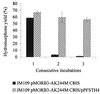
We have applied the soluble pyridine nucleotide transhydrogenase of Pseudomonas fluorescens to a cell-free system for the regeneration of the nicotinamide cofactors NAD and NADP in the biological production of the important semisynthetic opiate drug hydromorphone. The original recombinant whole-cell system suffered from cofactor depletion resulting from the action of an NADP(+)-dependent morphine dehydrogenase and an NADH-dependent morphinone reductase. By applying a soluble pyridine nucleotide transhydrogenase, which can transfer reducing equivalents between NAD and NADP, we demonstrate with a cell-free system that efficient cofactor cycling in the presence of catalytic amounts of cofactors occurs, resulting in high yields of hydromorphone. The ratio of morphine dehydrogenase, morphinone reductase, and soluble pyridine nucleotide transhydrogenase is critical for diminishing the production of the unwanted by-product dihydromorphine and for optimum hydromorphone yields. Application of the soluble pyridine nucleotide transhydrogenase to the whole-cell system resulted in an improved biocatalyst with an extended lifetime. These results demonstrate the usefulness of the soluble pyridine nucleotide transhydrogenase and its wider application as a tool in metabolic engineering and biocatalysis.
Figures




References
-
- Anderlund M, Nissen T L, Nielsen J, Villadsen J, Rydström J, HahnHagerdal B, KiellandBrandt M C. Expression of the Escherichia coli pntA and pntB genes, encoding nicotinamide nucleotide transhydrogenase, in Saccharomyces cerevisiae and its effect on product formation during anaerobic glucose fermentation. Appl Environ Microbiol. 1999;65:2333–2340. - PMC - PubMed
-
- Andersen K B, von Meyenburg K. Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J Biol Chem. 1977;252:4151–4156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

