Control of expression of divergent Pseudomonas putida put promoters for proline catabolism
- PMID: 11097893
- PMCID: PMC92447
- DOI: 10.1128/AEM.66.12.5221-5225.2000
Control of expression of divergent Pseudomonas putida put promoters for proline catabolism
Abstract
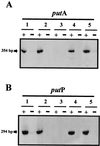
Pseudomonas putida KT2440 uses proline as the sole C and N source. Utilization of this amino acid involves its uptake, which is mediated by the PutP protein, and its conversion into glutamate, mediated by the PutA protein. Sequence analysis revealed that the putA and putP genes are transcribed divergently. Expression from the putP and putA genes was analyzed at the mRNA level in different host backgrounds in the absence and presence of proline. Expression from the put promoters was induced by proline. The transcription initiation points of the putP and putA genes were precisely mapped via primer extension, and sequence analysis of the upstream DNA region showed well-separated promoters for these two genes. The PutA protein acts as a repressor of put gene expression in P. putida because expression from the put promoters is constitutive in a host background with a knockout putA gene. This regulatory activity is independent of the catabolic activity of PutA, because we show that a point mutation (Glu896-->Lys) that prevents catalytic activity allowed the protein to retain its regulatory activity. Expression from the put promoters in the presence of proline in a putA-proficient background requires a positive regulatory protein, still unidentified, whose expression seems to be sigma(54) dependent because the put genes were not expressed in a sigma(54)-deficient background. Expression of the putA and putP genes was equally high in the presence of proline in sigma(38)- and ihf-deficient P. putida backgrounds.
Figures



References
-
- Brown E D, Wood J M. Conformational change and membrane association of the PutA protein are coincident with reduction of its FAD cofactor by proline. J Biol Chem. 1993;268:8972–8979. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

