PCR detection of Salmonella enterica serotype Montevideo in and on raw tomatoes using primers derived from hilA
- PMID: 11097898
- PMCID: PMC92452
- DOI: 10.1128/AEM.66.12.5248-5252.2000
PCR detection of Salmonella enterica serotype Montevideo in and on raw tomatoes using primers derived from hilA
Abstract
Salmonellae have been some of the most frequently reported etiological agents in fresh-produce-associated outbreaks of human infections in recent years. PCR assays using four innovative pairs of primers derived from hilA and sirA, positive regulators of Salmonella invasive genes, were developed to identify Salmonella enterica serotype Montevideo on and in tomatoes. Based on examination of 83 Salmonella strains and 22 non-Salmonella strains, we concluded that a pair of hilA primers detects Salmonella specifically. The detection limits of the PCR assay were 10(1) and 10(0) CFU/ml after enrichment at 37 degrees C for 6 and 9 h, respectively. When the assay was validated by detecting S. enterica serotype Montevideo in and on artificially inoculated tomatoes, 10(2) and 10(1) CFU/g were detected, respectively, after enrichment for 6 h at 37 degrees C. Our results suggest that the hilA-based PCR assay is sensitive and specific, and can be used for rapid detection of Salmonellae in or on fresh produce.
Figures

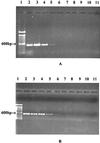

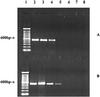
References
-
- Ahmer B M M, Heffron F. Salmonella typhimurium recognition of intestinal environments: response. Trends Microbiol. 1999;7:222–223. - PubMed
-
- Ahmer B M M, Van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. - PubMed
-
- Asplund K, Nurmi E. The growth of salmonellae in tomatoes. Int J Food Microbiol. 1991;13:177–182. - PubMed
-
- Bailey J S. Detection of Salmonella cells within 24–26 hrs in poultry samples with the PCR BAX system. J Food Prot. 1998;61:792–795. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

