Sequencing bands of ribosomal intergenic spacer analysis fingerprints for characterization and microscale distribution of soil bacterium populations responding to mercury spiking
- PMID: 11097911
- PMCID: PMC92465
- DOI: 10.1128/AEM.66.12.5334-5339.2000
Sequencing bands of ribosomal intergenic spacer analysis fingerprints for characterization and microscale distribution of soil bacterium populations responding to mercury spiking
Abstract
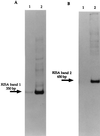
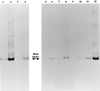
Two major emerging bands (a 350-bp band and a 650-bp band) within the RISA (ribosomal intergenic spacer analysis) profile of a soil bacterial community spiked with Hg(II) were selected for further identification of the populations involved in the response of the community to the added metal. The bands were cut out from polyacrylamide gels, cloned, characterized by restriction analysis, and sequenced for phylogenetic affiliation of dominant clones. The sequences were the intergenic spacer between the rrs and rrl genes and the first 130 nucleotides of the rrl gene. Comparison of sequences derived from the 350-bp band to The GenBank database permitted us to identify the bacteria as being mostly close relatives to low G+C firmicutes (Clostridium-like genera), while the 650-bp band permitted us to identify the bacteria as being mostly close relatives to beta-proteobacteria (Ralstonia-like genera). Oligonucleotide probes specific for the identified dominant bacteria were designed and hybridized with the RISA profiles derived from the control and spiked communities. These studies confirmed the contribution of these populations to the community response to the metal. Hybridization of the RISA profiles from subcommunities (bacterial pools associated with different soil microenvironments) also permitted to characterize the distribution and the dynamics of these populations at a microscale level following mercury spiking.
Figures

References
-
- Barkay T, Turner R R, Vandenbrook A, Liebert C. The relationships of Hg(II) volatilization from a freshwater pond to the abundance of mer genes in the gene pool of the indigenous microbial community. Microb Ecol. 1991;21:151–161. - PubMed
-
- Barbieri P, Bestetti G, Reniero D, Galli E. Mercury resistance in aromatic compound degrading Pseudomonas strains. FEMS Microbiol Ecol. 1996;62:375–384.
-
- Barry T, Colleran G, Glennon M, Dunican L K, Gannon F. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991;1:51–56. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

