Isolation, purification, and characterization of a killer protein from Schwanniomyces occidentalis
- PMID: 11097913
- PMCID: PMC92467
- DOI: 10.1128/AEM.66.12.5348-5352.2000
Isolation, purification, and characterization of a killer protein from Schwanniomyces occidentalis
Abstract
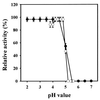
The yeast Schwanniomyces occidentalis produces a killer toxin lethal to sensitive strains of Saccharomyces cerevisiae. Killer activity is lost after pepsin and papain treatment, suggesting that the toxin is a protein. We purified the killer protein and found that it was composed of two subunits with molecular masses of approximately 7.4 and 4.9 kDa, respectively, but was not detectable with periodic acid-Schiff staining. A BLAST search revealed that residues 3 to 14 of the 4.9-kDa subunit had 75% identity and 83% similarity with killer toxin K2 from S. cerevisiae at positions 271 to 283. Maximum killer activity was between pH 4.2 and 4.8. The protein was stable between pH 2.0 and 5.0 and inactivated at temperatures above 40 degrees C. The killer protein was chromosomally encoded. Mannan, but not beta-glucan or laminarin, prevented sensitive yeast cells from being killed by the killer protein, suggesting that mannan may bind to the killer protein. Identification and characterization of a killer strain of S. occidentalis may help reduce the risk of contamination by undesirable yeast strains during commercial fermentations.
Figures


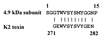

Similar articles
-
Investigation of a killer strain of Zygosaccharomyces bailii.J Gen Microbiol. 1993 Mar;139(3):495-500. doi: 10.1099/00221287-139-3-495. J Gen Microbiol. 1993. PMID: 8473858
-
Characterization of a novel killer toxin encoded by a double-stranded linear DNA plasmid of Kluyveromyces lactis.Eur J Biochem. 1984 Jun 1;141(2):241-5. doi: 10.1111/j.1432-1033.1984.tb08183.x. Eur J Biochem. 1984. PMID: 6734597
-
The killer toxin of Kluyveromyces lactis: characterization of the toxin subunits and identification of the genes which encode them.EMBO J. 1986 Aug;5(8):1995-2002. doi: 10.1002/j.1460-2075.1986.tb04455.x. EMBO J. 1986. PMID: 3758030 Free PMC article.
-
Genetic and molecular approaches to synthesis and action of the yeast killer toxin.Experientia. 1990 Feb 15;46(2):193-200. doi: 10.1007/BF02027313. Experientia. 1990. PMID: 2406163 Review.
-
K1 killer toxin, a pore-forming protein from yeast.Mol Microbiol. 1991 Oct;5(10):2339-43. doi: 10.1111/j.1365-2958.1991.tb02079.x. Mol Microbiol. 1991. PMID: 1724277 Review.
Cited by
-
Killer toxin of Kluyveromyces phaffii DBVPG 6076 as a biopreservative agent to control apiculate wine Yeasts.Appl Environ Microbiol. 2001 Jul;67(7):3058-63. doi: 10.1128/AEM.67.7.3058-3063.2001. Appl Environ Microbiol. 2001. PMID: 11425722 Free PMC article.
-
Susceptibility of Candida albicans Isolated from Blood to Wickerhamomyces anomalous Mycocins.Curr Microbiol. 2016 Dec;73(6):878-884. doi: 10.1007/s00284-016-1135-4. Epub 2016 Sep 16. Curr Microbiol. 2016. PMID: 27638312
-
Diversity of fermentative yeasts with probiotic potential isolated from Thai fermented food products.AIMS Microbiol. 2022 Dec 26;8(4):575-594. doi: 10.3934/microbiol.2022037. eCollection 2022. AIMS Microbiol. 2022. PMID: 36694589 Free PMC article.
-
Antimicrobial and probiotic properties of yeasts: from fundamental to novel applications.Front Microbiol. 2012 Dec 19;3:421. doi: 10.3389/fmicb.2012.00421. eCollection 2012. Front Microbiol. 2012. PMID: 23267352 Free PMC article.
-
Identification of additional probiotic attributes in yeasts isolated from tarhana fermentation.Braz J Microbiol. 2025 Sep;56(3):1765-1773. doi: 10.1007/s42770-025-01728-4. Epub 2025 Jul 9. Braz J Microbiol. 2025. PMID: 40632463
References
-
- Bevan E A, Makower M. Proceedings of the XIth International Congress on Genetics. 1963. The physiological basis of the killer character in yeast; pp. 202–203.
-
- Bollag D M, Rozycki M D, Edelstein S J. Gel electrophoresis under nondenaturing conditions. In: Bollag D M, Rozycki M D, Edelstein S J, editors. Protein methods. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 155–172.
-
- Bortol A, Nudel C, Fraille E, De Torres R, Giuletti A, Spencer J F T, Spencer D. Isolation of yeast with killer activity and its breeding with an industrial baking strain by protoplast fusion. Appl Microbiol Biotechnol. 1986;24:414–416.
-
- Bostian K A, Hopper J E, Rogers D T, Tipper D J. Translational analysis of the killer-associated virus-like particle dsRNA genome of S. cerevisiae: M dsRNA encodes toxin. Cell. 1980;19:403–414. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

