Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil
- PMID: 11097926
- PMCID: PMC92480
- DOI: 10.1128/AEM.66.12.5448-5456.2000
Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil
Abstract
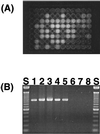
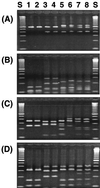
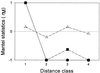
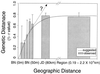
Fluorescent Pseudomonas strains were isolated from 38 undisturbed pristine soil samples from 10 sites on four continents. A total of 248 isolates were confirmed as Pseudomonas sensu stricto by fluorescent pigment production and group-specific 16S ribosomal DNA (rDNA) primers. These isolates were analyzed by three molecular typing methods with different levels of resolution: 16S rDNA restriction analysis (ARDRA), 16S-23S rDNA intergenic spacer-restriction fragment length polymorphism (ITS-RFLP) analysis, and repetitive extragenic palindromic PCR genomic fingerprinting with a BOX primer set (BOX-PCR). All isolates showed very similar ARDRA patterns, as expected. Some ITS-RFLP types were also found at every geographic scale, although some ITS-RFLP types were unique to the site of origin, indicating weak endemicity at this level of resolution. Using a similarity value of 0.8 or more after cluster analysis of BOX-PCR fingerprinting patterns to define the same genotypes, we identified 85 unique fluorescent Pseudomonas genotypes in our collection. There were no overlapping genotypes between sites as well as continental regions, indicating strict site endemism. The genetic distance between isolates as determined by degree of dissimilarity in BOX-PCR patterns was meaningfully correlated to the geographic distance between the isolates' sites of origin. Also, a significant positive spatial autocorrelation of the distribution of the genotypes was observed among distances of <197 km, and significant negative autocorrelation was observed between regions. Hence, strong endemicity of fluorescent Pseudomonas genotypes was observed, suggesting that these heterotrophic soil bacteria are not globally mixed.
Figures


References
-
- Baas-Becking L G M. Geobiologie of Inleiding Tot de Milieukunde. The Hague, The Netherlands: W. P. van Stockum & Zoon N. V.; 1934.
-
- Beijerinck M W. De infusies en de ontdekking der backteriën, Jaarboek van de Koninklijke Akademie v. Wetenschappen. Amsterdam, The Netherlands: Müller; 1913.
-
- Castenholz R W. Endemism and biodiversity of thermophilic cyanobacteria. Nova Hedwigia Beiheft. 1996;112:33–47.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

