Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the "Bacillus cereus group"
- PMID: 11097928
- PMCID: PMC92482
- DOI: 10.1128/AEM.66.12.5460-5468.2000
Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the "Bacillus cereus group"
Abstract
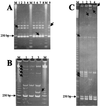
Bacillus anthracis, Bacillus cereus, Bacillus mycoides, Bacillus pseudomycoides, Bacillus thuringiensis, and Bacillus weihenstephanensis are closely related in phenotype and genotype, and their genetic relationship is still open to debate. The present work uses amplified 16S-23S internal transcribed spacers (ITS) to discriminate between the strains and species and to describe the genetic relationships within the "B. cereus group," advantage being taken of homoduplex-heteroduplex polymorphisms (HHP) resolved by polyacrylamide gel electrophoresis and silver staining. One hundred forty-one strains belonging to the six species were investigated, and 73 ITS-HHP pattern types were distinguished by MDE, a polyacrylamide matrix specifically designed to resolve heteroduplex and single-strand conformation polymorphisms. The discriminating bands were confirmed as ITS by Southern hybridization, and the homoduplex or heteroduplex nature was identified by single-stranded DNA mung bean nuclease digestion. Several of the ITS-HHP types corresponded to specific phenotypes such as B. anthracis or serotypes of B. thuringiensis. Unweighted pair group method arithmetic average cluster analysis revealed two main groups. One included B. mycoides, B. weihenstephanensis, and B. pseudomycoides. The second included B. cereus and B. thuringiensis, B. anthracis appeared as a lineage of B. cereus.
Figures


References
-
- Andreoni V, Moro Luischi M, Cavalca L, Erba D, Ciapellano S. Selenite tolerance and accumulation in the Lactobacillus species. Ann Microbiol. 2000;50:77–88.
-
- Ash C, Collins M D. Comparative analysis of 23S ribosomal RNA gene sequences of Bacillus anthracis and emetic Bacillus cereus determined by PCR-direct sequencing. FEMS Microbiol Lett. 1992;94:75–80. - PubMed
-
- Ash C, Farrow J A E, Dorsch M, Stackebrandt E, Collins M D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. - PubMed
-
- Ash C, Farrow J A E, Wallbanks S, Collins M D. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett Appl Microbiol. 1991;13:202–206.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

