Autophagy as a regulated pathway of cellular degradation
- PMID: 11099404
- PMCID: PMC2732363
- DOI: 10.1126/science.290.5497.1717
Autophagy as a regulated pathway of cellular degradation
Abstract
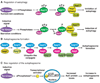
Macroautophagy is a dynamic process involving the rearrangement of subcellular membranes to sequester cytoplasm and organelles for delivery to the lysosome or vacuole where the sequestered cargo is degraded and recycled. This process takes place in all eukaryotic cells. It is highly regulated through the action of various kinases, phosphatases, and guanosine triphosphatases (GTPases). The core protein machinery that is necessary to drive formation and consumption of intermediates in the macroautophagy pathway includes a ubiquitin-like protein conjugation system and a protein complex that directs membrane docking and fusion at the lysosome or vacuole. Macroautophagy plays an important role in developmental processes, human disease, and cellular response to nutrient deprivation.
Figures

References
-
- Mortimore GE, Miotto G, Venerando R, Kadowaki M. Subcell. Biochem. 1996;27:93. - PubMed
-
- Vittorini S, et al. J. Gerontol. A Biol. Sci. Med. Sci. 1999;54:B318. - PubMed
-
- Dunn WA., Jr Trends Cell Biol. 1994;4:139. - PubMed
-
- Cuervo AM, Dice JF. J. Mol. Med. 1998;76:6. - PubMed
-
- Kim J, Klionsky DJ. Annu. Rev. Biochem. 2000;69:303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

