The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view
- PMID: 11101504
- PMCID: PMC305862
- DOI: 10.1093/emboj/19.23.6317
The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view
Abstract
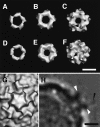
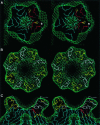
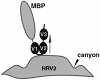
Human rhinovirus serotype 2 (HRV2) belongs to the minor group of HRVs that bind to members of the LDL-receptor family including the very low density lipoprotein (VLDL)-receptor (VLDL-R). We have determined the structures of the complex between HRV2 and soluble fragments of the VLDL-R to 15 A resolution by cryo-electron microscopy. The receptor fragments, which include the first three ligand-binding repeats of the VLDL-R (V1-3), bind to the small star-shaped dome on the icosahedral 5-fold axis. This is in sharp contrast to the major group of HRVs where the receptor site for ICAM-1 is located at the base of a depression around each 5-fold axis. Homology models of the three domains of V1-3 were used to explore the virus-receptor interaction. The footprint of VLDL-R on the viral surface covers the BC- and HI-loops on VP1.
Figures



Similar articles
-
A cellular receptor of human rhinovirus type 2, the very-low-density lipoprotein receptor, binds to two neighboring proteins of the viral capsid.J Virol. 2003 Aug;77(15):8504-11. doi: 10.1128/jvi.77.15.8504-8511.2003. J Virol. 2003. PMID: 12857919 Free PMC article.
-
X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein.Nat Struct Mol Biol. 2004 May;11(5):429-34. doi: 10.1038/nsmb753. Epub 2004 Apr 4. Nat Struct Mol Biol. 2004. PMID: 15064754
-
Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor.EMBO J. 1999 Nov 15;18(22):6249-59. doi: 10.1093/emboj/18.22.6249. EMBO J. 1999. PMID: 10562537 Free PMC article.
-
Review: rhinoviruses and their ICAM receptors.J Struct Biol. 1999 Dec 1;128(1):69-74. doi: 10.1006/jsbi.1999.4143. J Struct Biol. 1999. PMID: 10600561 Review.
-
ICAM-1 receptors and cold viruses.Pharm Acta Helv. 2000 Mar;74(2-3):291-7. doi: 10.1016/s0031-6865(99)00056-4. Pharm Acta Helv. 2000. PMID: 10812972 Free PMC article. Review.
Cited by
-
Nonneutralizing human rhinovirus serotype 2-specific monoclonal antibody 2G2 attaches to the region that undergoes the most dramatic changes upon release of the viral RNA.J Virol. 2006 Dec;80(24):12398-401. doi: 10.1128/JVI.01399-06. Epub 2006 Sep 27. J Virol. 2006. PMID: 17005641 Free PMC article.
-
Human rhinovirus subviral a particle binds to lipid membranes over a twofold axis of icosahedral symmetry.J Virol. 2013 Oct;87(20):11309-12. doi: 10.1128/JVI.02055-13. Epub 2013 Aug 14. J Virol. 2013. PMID: 23946453 Free PMC article.
-
Population structure and evolution of Rhinoviruses.PLoS One. 2014 Feb 19;9(2):e88981. doi: 10.1371/journal.pone.0088981. eCollection 2014. PLoS One. 2014. PMID: 24586469 Free PMC article.
-
Modeling of the human rhinovirus C capsid suggests a novel topography with insights on receptor preference and immunogenicity.Virology. 2014 Jan 5;448:176-84. doi: 10.1016/j.virol.2013.10.006. Epub 2013 Oct 25. Virology. 2014. PMID: 24314648 Free PMC article.
-
Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody.Nat Commun. 2017 Sep 11;8(1):505. doi: 10.1038/s41467-017-00477-9. Nat Commun. 2017. PMID: 28894095 Free PMC article.
References
-
- Appleyard G., Russell,S.M., Clarke,B.E., Speller,S.A., Trowbridge,M. and Vadolas,J. (1990) Neutralization epitopes of human rhinovirus type 2. J. Gen. Virol., 71, 1275–1282. - PubMed
-
- Baker T.S. and Cheng,R.H. (1996) A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol., 116, 120–130. - PubMed
-
- Brown M.S. and Goldstein,J.L. (1986) A receptor-mediated pathway for cholesterol homeostasis. Science, 232, 34–47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

