Nud1p links astral microtubule organization and the control of exit from mitosis
- PMID: 11101520
- PMCID: PMC305870
- DOI: 10.1093/emboj/19.23.6475
Nud1p links astral microtubule organization and the control of exit from mitosis
Erratum in
- EMBO J 2001 Jan 15;20(1-2):305
Abstract
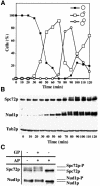
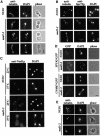
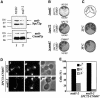
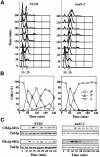
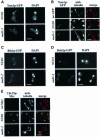
The budding yeast spindle pole body (SPB) not only organizes the astral and nuclear microtubules but is also associated with a number of cell-cycle regulators that control mitotic exit. Here, we describe that the core SPB component Nud1p is a key protein that functions in both processes. The astral microtubule organizing function of Nud1p is mediated by its interaction with the gamma-tubulin complex binding protein Spc72p. This function of Nud1p is distinct from its role in cell-cycle control: Nud1p binds the spindle checkpoint control proteins Bfa1p and Bub2p to the SPB, and is part of the mitotic exit network (MEN) in which it functions upstream of CDC15 but downstream of LTE1. In conditional lethal nud1-2 cells, the MEN component Tem1p, a GTPase, is mislocalized, whereas the kinase Cdc15p is still associated with the SPB. Thus, in nud1-2 cells the failure of Tem1p to interact with Cdc15p at the SPB probably prevents mitotic exit.
Figures




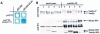
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

