Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome
- PMID: 11101533
- PMCID: PMC305868
- DOI: 10.1093/emboj/19.23.6612
Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome
Abstract
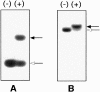
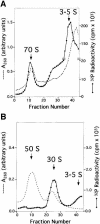
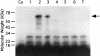
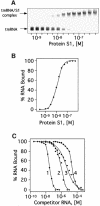
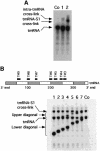
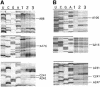
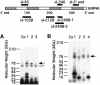
UV irradiation of an in vitro translation mixture induced cross-linking of 4-thioU-substituted tmRNA to Escherichia coli ribosomes by forming covalent complexes with ribosomal protein S1 and 16S rRNA. In the absence of S1, tmRNA was unable to bind and label ribosomal components. Mobility assays on native gels demonstrated that protein S1 bound to tmRNA with an apparent binding constant of 1 x 10(8) M(-1). A mutant tmRNA, lacking the tag coding region and pseudoknots pk2, pk3 and pk4, did not compete with full-length tmRNA, indicating that this region is required for S1 binding. This was confirmed by identification of eight cross-linked nucleotides: U85, located before the resume codon of tmRNA; U105, in the mRNA portion of tmRNA; U172 in pK2; U198, U212, U230 and U240 in pk3; and U246, in the junction between pk3 and pk4. We concluded that ribosomal protein S1, in concert with the previously identified elongation factor EF-Tu and protein SmpB, plays an important role in tmRNA-mediated trans-translation by facilitating the binding of tmRNA to ribosomes and forming complexes with free tmRNA.
Figures

References
-
- Barends S., Wower,J. and Kraal,B. (2000) Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry, 39, 2652–2658. - PubMed
-
- Barrera I., Schuppli,D., Sogo,J.M. and Weber H. (1993) Different mechanisms of recognition of bacteriophage Qβ plus and minus strand RNAs by Qβ replicase. J. Mol. Biol., 232, 512–521. - PubMed
-
- Berkower I., Leis,J. and Hurwitz,J. (1973) Isolation and characterization of an endonuclease from Escherichia coli specific for ribonucleic acid in ribonucleic acid–deoxyribonucleic acid hybrid structures. J. Biol. Chem., 248, 5914–5921. - PubMed
-
- Brimacombe R., Greuer,B., Gulle,H., Kosack,M., Mitchell,P., Osswald,M., Stade,K. and Stiege,W. (1990) New techniques for the analysis of intra-RNA and RNA–protein cross-linking data from ribosomes. In Spedding,G. (ed.), Ribosomes and Protein Synthesis. A Practical Approach. Oxford University Press, Oxford, UK, pp. 131–159.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

