Genomic and phylogenetic analysis of hepatitis C virus isolates from argentine patients: a six-year retrospective study
- PMID: 11101596
- PMCID: PMC87637
- DOI: 10.1128/JCM.38.12.4560-4568.2000
Genomic and phylogenetic analysis of hepatitis C virus isolates from argentine patients: a six-year retrospective study
Erratum in
- J Clin Microbiol 2001 Mar;39(3):1208-10
Abstract
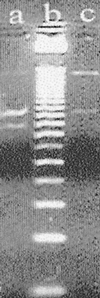
Typing of hepatitis C virus (HCV) isolates from Argentine patients was performed by using different methodologies in a population of 243 patients. HCV subtype was assigned based upon restriction fragment length polymorphism (RFLP). HCV RNA genomes obtained from serum samples were classified as belonging to clade 1 (53.5%), 2 (23. 0%), or 3 (8.6%); 14.8% of samples showed HCV mixed infections, more frequently implying different subtypes within the same clade. In addition to RFLP typing, phylogenetic relatedness among sequences from both 5' untranslated region (n = 50) and nonstructural 5B coding region (n = 15) was established.
Figures
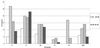


References
-
- Abrignani S, Rosa D. Perspectives for a hepatitis C virus vaccine. Clin Diagn Virol. 1998;15:181–185. - PubMed
-
- Buoro S, Pizzighella S, Boschetto R, Pellizzari L, Cusan M, Bonaguro R, Mengoli C, Caudai C, Padula M, Valensin P E, Palù G. Typing of hepatitis C virus by a new method based on restriction fragment length polymorphism. Intervirology. 1999;42:1–8. - PubMed
-
- Castillo I, Bartolome J, Quiroga J A, Carreño V. Comparison of several PCR procedures for detection of serum HCV-RNA using different regions of the HCV genome. J Virol Methods. 1992;38:71–79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

