GABAC receptor sensitivity is modulated by interaction with MAP1B
- PMID: 11102469
- PMCID: PMC6773065
- DOI: 10.1523/JNEUROSCI.20-23-08643.2000
GABAC receptor sensitivity is modulated by interaction with MAP1B
Abstract
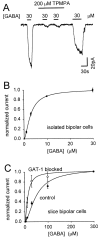
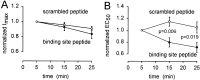
GABA(C) receptors contain rho subunits and mediate feedback inhibition from retinal amacrine cells to bipolar cells. We previously identified the cytoskeletal protein MAP1B as a rho1 subunit anchoring protein. Here, we analyze the structural basis and functional significance of the MAP1B-rho1 interaction. Twelve amino acids at the C terminus of the large intracellular loop of rho1 (and also rho2) are sufficient for interaction with MAP1B. Disruption of the MAP1B-rho interaction in bipolar cells in retinal slices decreased the EC(50) of their GABA(C) receptors, doubling the receptors' current at low GABA concentrations without affecting their maximum current at high concentrations. Thus, anchoring to the cytoskeleton lowers the sensitivity of GABA(C) receptors and provides a likely site for functional modulation of GABA(C) receptor-mediated inhibition.
Figures





References
-
- Borden LA, Dhar TGM, Smith KE, Weinshank RL, Branchek TA, Gluchowski C. Tiagabine, SK&F 89976-A, CI-966 and NNC-711 are selective for the cloned GABA transporter GAT-1. Eur J Pharmacol. 1994;269:219–224. - PubMed
-
- Calvo DJ, Miledi R. Activation of GABAρ1 receptors by glycine and β-alanine. NeuroReport. 1995;6:1118–1120. - PubMed
-
- Chang Y, Weiss DS. Channel opening locks agonist onto the GABAC receptor. Nat Neurosci. 1999;2:219–225. - PubMed
-
- Cutting GR, Lu L, O'Hara BF, Kasch LM, Montrose-Rafizadeh C, Donovan DM, Shimada S, Antonarakis SE, Guggino WB, Uhl GR, Kazazian HH., Jr Cloning of the γ-aminobutyric acid (GABA) ρ1 cDNA: a GABA receptor subunit highly expressed in the retina. Proc Natl Acad Sci USA. 1991;88:2673–2677. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
