Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes
- PMID: 11102511
- PMCID: PMC15060
- DOI: 10.1091/mbc.11.12.4105
Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes
Abstract
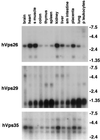
Sorting nexin (SNX) 1 and SNX2 are mammalian orthologs of Vps5p, a yeast protein that is a subunit of a large multimeric complex, termed the retromer complex, involved in retrograde transport of proteins from endosomes to the trans-Golgi network. We report the cloning and characterization of human orthologs of three additional components of the complex: Vps26p, Vps29p, and Vps35p. The close structural similarity between the yeast and human proteins suggests a similarity in function. We used both yeast two-hybrid assays and expression in mammalian cells to define the binding interactions among these proteins. The data suggest a model in which hVps35 serves as the core of a multimeric complex by binding directly to hVps26, hVps29, and SNX1. Deletional analyses of hVps35 demonstrate that amino acid residues 1-53 and 307-796 of hVps35 bind to the coiled coil-containing domain of SNX1. In contrast, hVps26 binds to amino acid residues 1-172 of hVps35, whereas hVps29 binds to amino acid residues 307-796 of hVps35. Furthermore, hVps35, hVps29, and hVps26 have been found in membrane-associated and cytosolic compartments. Gel filtration chromatography of COS7 cell cytosol showed that both recombinant and endogenous hVps35, hVps29, and hVps26 coelute as a large complex ( approximately 220-440 kDa). In the absence of hVps35, neither hVps26 nor hVps29 is found in the large complex. These data provide the first insights into the binding interactions among subunits of a putative mammalian retromer complex.
Figures











References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Vol. 2. New York: John Wiley & Sons; 1994. pp. 13.17.11–13.17.10.
-
- Bachhawat AK, Suhan J, Jones EW. The yeast homolog of H 58, a mouse gene essential for embryogenesis, performs a role in the delivery of proteins to the vacuole. Genes Dev. 1994;8:1379–1387. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

