Mutation of cyclin/cdk phosphorylation sites in HsCdc6 disrupts a late step in initiation of DNA replication in human cells
- PMID: 11102512
- PMCID: PMC15061
- DOI: 10.1091/mbc.11.12.4117
Mutation of cyclin/cdk phosphorylation sites in HsCdc6 disrupts a late step in initiation of DNA replication in human cells
Abstract
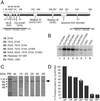
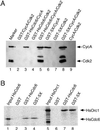
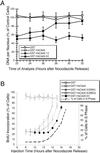
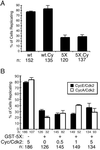
Cyclin-dependent kinases (Cdk) are essential for promoting the initiation of DNA replication, presumably by phosphorylating key regulatory proteins that are involved in triggering the G1/S transition. Human Cdc6 (HsCdc6), a protein required for initiation of DNA replication, is phosphorylated by Cdk in vitro and in vivo. Here we report that HsCdc6 with mutations at potential Cdk phosphorylation sites was poorly phosphorylated in vitro by Cdk, but retained all other biochemical activities of the wild-type protein tested. Microinjection of mutant HsCdc6 proteins into human cells blocked initiation of DNA replication or slowed S phase progression. The inhibitory effect of mutant HsCdc6 was lost at the G1/S transition, indicating that phosphorylation of HsCdc6 by Cdk is critical for a late step in initiation of DNA replication in human cells.
Figures

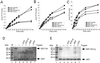

References
-
- Aparicio OM, Weinstein DM, Bell. SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Araki T, Yamamoto A, Yamada M. Accurate determination of DNA content in single cell nuclei stained with Hoechst 33258 fluorochrome at high salt concentration. Histochemistry. 1987;87:331–338. - PubMed
-
- Calzada A, Sánchez M, Sánchez E, Bueno A. The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J Biol Chem. 2000;275:9734–9741. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

