A role for poly(ADP-ribose) polymerase in the transcriptional regulation of the melanoma growth stimulatory activity (CXCL1) gene expression
- PMID: 11112786
- PMCID: PMC3369623
- DOI: 10.1074/jbc.M009897200
A role for poly(ADP-ribose) polymerase in the transcriptional regulation of the melanoma growth stimulatory activity (CXCL1) gene expression
Abstract
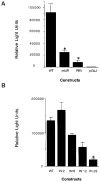
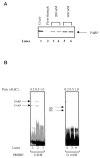
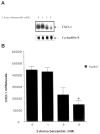
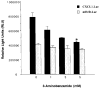
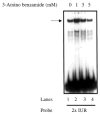
The melanoma growth stimulatory activity/growth-regulated protein, CXCL1, is constitutively expressed at high levels during inflammation and progression of melanocytes into malignant melanoma. It has been shown previously that CXCL1 overexpression in melanoma cells is due to increased transcription as well as stability of the CXCL1 message. The transcription of CXCL1 is regulated through several cis-acting elements including Sp1, NF-kappaB, HMGI(Y), and the immediate upstream region (IUR) element (nucleotides -94 to -78), which lies immediately upstream to the nuclear factor kappaB (NF-kappaB) element. Previously, it has been shown that the IUR is necessary for basal and cytokine-induced transcription of the CXCL1 gene. UV cross-linking and Southwestern blot analyses indicate that the IUR oligonucleotide probe selectively binds a 115-kDa protein. In this study, the IUR element has been further characterized. We show here that proximity of the IUR element to the adjacent NF-kappaB element is critical to its function as a positive regulatory element. Using binding site oligonucleotide affinity chromatography, we have selectively purified the 115-kDa IUR-F. Mass spectrometry/mass spectrometry/matrix-assisted laser desorption ionization/time of flight spectroscopy and amino acid analysis as well as microcapillary reverse phase chromatography electrospray ionization tandem mass spectrometry identified this protein as the 114-kDa poly(ADP-ribose) polymerase (PARP1). Furthermore, 3-aminobenzamide, an inhibitor of PARP-specific ADP-ribosylation, inhibits CXCL1 promoter activity and reduces levels of CXCL1 mRNA. The data point to the possibility that PARP may be a coactivator of CXCL1 transcription.
Figures


Similar articles
-
Differential regulation of CXC ligand 1 transcription in melanoma cell lines by poly(ADP-ribose) polymerase-1.Oncogene. 2006 Dec 14;25(59):7714-22. doi: 10.1038/sj.onc.1209751. Epub 2006 Jun 26. Oncogene. 2006. PMID: 16799643 Free PMC article.
-
Constitutive and cytokine-induced expression of the melanoma growth stimulatory activity/GRO alpha gene requires both NF-kappa B and novel constitutive factors.J Biol Chem. 1995 Dec 22;270(51):30619-26. doi: 10.1074/jbc.270.51.30619. J Biol Chem. 1995. PMID: 8530498
-
HMGI(Y) and Sp1 in addition to NF-kappa B regulate transcription of the MGSA/GRO alpha gene.Nucleic Acids Res. 1995 Oct 25;23(20):4210-9. doi: 10.1093/nar/23.20.4210. Nucleic Acids Res. 1995. PMID: 7479086 Free PMC article.
-
Role of CXCL1 in tumorigenesis of melanoma.J Leukoc Biol. 2002 Jul;72(1):9-18. J Leukoc Biol. 2002. PMID: 12101257 Free PMC article. Review.
-
Mechanism and biological significance of constitutive expression of MGSA/GRO chemokines in malignant melanoma tumor progression.J Leukoc Biol. 1997 Nov;62(5):588-97. doi: 10.1002/jlb.62.5.588. J Leukoc Biol. 1997. PMID: 9365113 Review.
Cited by
-
Okamoto model for necrosis and its expansions, CD38-cyclic ADP-ribose signal system for intracellular Ca2+ mobilization and Reg (Regenerating gene protein)-Reg receptor system for cell regeneration.Proc Jpn Acad Ser B Phys Biol Sci. 2021;97(8):423-461. doi: 10.2183/pjab.97.022. Proc Jpn Acad Ser B Phys Biol Sci. 2021. PMID: 34629354 Free PMC article.
-
Poly(ADP-ribose) polymerase-1 (Parp-1)-deficient mice demonstrate abnormal antibody responses.Immunology. 2009 Jun;127(2):178-86. doi: 10.1111/j.1365-2567.2008.02921.x. Epub 2008 Sep 4. Immunology. 2009. PMID: 18778284 Free PMC article.
-
PARP1 as a Marker of an Aggressive Clinical Phenotype in Cutaneous Melanoma-A Clinical and an In Vitro Study.Cells. 2021 Jan 31;10(2):286. doi: 10.3390/cells10020286. Cells. 2021. PMID: 33572647 Free PMC article.
-
Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation.Curr Opin Cell Biol. 2008 Jun;20(3):294-302. doi: 10.1016/j.ceb.2008.03.006. Epub 2008 Apr 29. Curr Opin Cell Biol. 2008. PMID: 18450439 Free PMC article. Review.
-
Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes.Mol Cell Biol. 2012 Jul;32(13):2490-502. doi: 10.1128/MCB.06667-11. Epub 2012 Apr 30. Mol Cell Biol. 2012. PMID: 22547677 Free PMC article.
References
-
- Richmond A, Shattuck RL. In: Chemoattractant Ligands and Their Receptors. Horuk R, editor. CRC Press; Boca Raton, FL: 1996. pp. 87–124.
-
- Wang XC, Jobin C, Allen JB, Roberts WL, Jaffe GJ. Invest Ophthal Vis Sci. 1999;40:477–486. - PubMed
-
- Shattuck-Brandt RL, Richmond A. Cancer Res. 1997;57:3032–3039. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

