Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae
- PMID: 11113188
- PMCID: PMC88787
- DOI: 10.1128/MCB.21.1.136-147.2001
Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae
Abstract
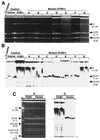
Saccharomyces cerevisiae carries approximately 150 ribosomal DNA (rDNA) copies in tandem repeats. Each repeat consists of the 35S rRNA gene, the NTS1 spacer, the 5S rRNA gene, and the NTS2 spacer. The FOB1 gene was previously shown to be required for replication fork block (RFB) activity at the RFB site in NTS1, for recombination hot spot (HOT1) activity, and for rDNA repeat expansion and contraction. We have constructed a strain in which the majority of rDNA repeats are deleted, leaving two copies of rDNA covering the 5S-NTS2-35S region and a single intact NTS1, and whose growth is supported by a helper plasmid carrying, in addition to the 5S rRNA gene, the 35S rRNA coding region fused to the GAL7 promoter. This strain carries a fob1 mutation, and an extensive expansion of chromosomal rDNA repeats was demonstrated by introducing the missing FOB1 gene by transformation. Mutational analysis using this system showed that not only the RFB site but also the adjacent approximately 400-bp region in NTS1 (together called the EXP region) are required for the FOB1-dependent repeat expansion. This approximately 400-bp DNA element is not required for the RFB activity or the HOT1 activity and therefore defines a function unique to rDNA repeat expansion (and presumably contraction) separate from HOT1 and RFB activities.
Figures
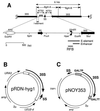
 ). (B) Structure of pRDN-hyg1 (4). This plasmid carries a single copy of rDNA repeats obtained by cutting the repeats with SmaI (hence the copy starting from −206 and ending at −207; the numbering is with respect to the Pol I transcription start site as +1). There is a mutation in the 18S rRNA gene (indicated as an asterisk) which makes the ribosome hygromycin B resistant. (C) Structure of pNOY353. This plasmid carries the 7.5-kb BamHI-XhoI fragment, which contains GAL7-35S rDNA (the 35S rRNA coding region fused to the GAL7 promoter as described by Nogi et al. [23]) inserted between the BamHI and SalI sites of the pTV3 vector (27). This plasmid also contains the 1,085-bp PvuII-EcoRV fragment carrying the 5S rRNA gene (see panel A) inserted in the SmaI site upstream of the GAL7 promoter. The 35S rRNA coding region contains up to the HindIII site, +6935. Thus, the Pol I enhancer is present but the region from HindIII to PvuII in NTS1 and the Pol I promoter region (from −1 to the EcoRV site at +8757 or the 381-bp region) are absent.
). (B) Structure of pRDN-hyg1 (4). This plasmid carries a single copy of rDNA repeats obtained by cutting the repeats with SmaI (hence the copy starting from −206 and ending at −207; the numbering is with respect to the Pol I transcription start site as +1). There is a mutation in the 18S rRNA gene (indicated as an asterisk) which makes the ribosome hygromycin B resistant. (C) Structure of pNOY353. This plasmid carries the 7.5-kb BamHI-XhoI fragment, which contains GAL7-35S rDNA (the 35S rRNA coding region fused to the GAL7 promoter as described by Nogi et al. [23]) inserted between the BamHI and SalI sites of the pTV3 vector (27). This plasmid also contains the 1,085-bp PvuII-EcoRV fragment carrying the 5S rRNA gene (see panel A) inserted in the SmaI site upstream of the GAL7 promoter. The 35S rRNA coding region contains up to the HindIII site, +6935. Thus, the Pol I enhancer is present but the region from HindIII to PvuII in NTS1 and the Pol I promoter region (from −1 to the EcoRV site at +8757 or the 381-bp region) are absent.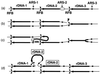
 , respectively. Individual lines represent chromatids with double-stranded DNA. In this model, DNA replication starts from one of the ARS sites (ARS-2) bidirectionally (a). In the yeast rDNA repeats, about one in five ARS sites is used as an active origin (2, 21). A rightward replication fork is arrested at the RFB site, and this arrest is supposed to stimulate a double-strand break of DNA at a nearby site (indicated by an arrowhead in row b). A strand invasion at a homologous duplex (a downstream sister chromatid near ARS-1 in this example) takes place (c), and a new replication fork is formed. The new replication fork meets with the leftward replication fork from the upstream site, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as boxed rDNA-2 (d). If the strand invasion is at a site in a upstream repeat (e.g., near ARS-3), a loss, rather than a gain, of an rDNA repeat is expected. This model was proposed previously to explain the observed strong dependence of rDNA repeat expansion and contraction on FOB1 (18).
, respectively. Individual lines represent chromatids with double-stranded DNA. In this model, DNA replication starts from one of the ARS sites (ARS-2) bidirectionally (a). In the yeast rDNA repeats, about one in five ARS sites is used as an active origin (2, 21). A rightward replication fork is arrested at the RFB site, and this arrest is supposed to stimulate a double-strand break of DNA at a nearby site (indicated by an arrowhead in row b). A strand invasion at a homologous duplex (a downstream sister chromatid near ARS-1 in this example) takes place (c), and a new replication fork is formed. The new replication fork meets with the leftward replication fork from the upstream site, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as boxed rDNA-2 (d). If the strand invasion is at a site in a upstream repeat (e.g., near ARS-3), a loss, rather than a gain, of an rDNA repeat is expected. This model was proposed previously to explain the observed strong dependence of rDNA repeat expansion and contraction on FOB1 (18).

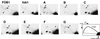
Similar articles
-
Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I.Genes Dev. 1998 Dec 15;12(24):3821-30. doi: 10.1101/gad.12.24.3821. Genes Dev. 1998. PMID: 9869636 Free PMC article.
-
Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae.Mol Cell Biol. 2006 Mar;26(6):2226-36. doi: 10.1128/MCB.26.6.2226-2236.2006. Mol Cell Biol. 2006. PMID: 16507999 Free PMC article.
-
Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae.Genes Cells. 2002 Feb;7(2):99-113. doi: 10.1046/j.1356-9597.2001.00508.x. Genes Cells. 2002. PMID: 11895475
-
[Recombination regulation by noncoding transcription in yeast rDNA repeats].Tanpakushitsu Kakusan Koso. 2006 Nov;51(14 Suppl):2141-3. Tanpakushitsu Kakusan Koso. 2006. PMID: 17471925 Review. Japanese. No abstract available.
-
Saccharomyces cerevisiae rDNA as super-hub: the region where replication, transcription and recombination meet.Cell Mol Life Sci. 2020 Dec;77(23):4787-4798. doi: 10.1007/s00018-020-03562-3. Epub 2020 May 31. Cell Mol Life Sci. 2020. PMID: 32476055 Free PMC article. Review.
Cited by
-
Natural genetic variation in yeast longevity.Genome Res. 2012 Oct;22(10):1963-73. doi: 10.1101/gr.136549.111. Epub 2012 Sep 5. Genome Res. 2012. PMID: 22955140 Free PMC article.
-
Coevolution of ribosomal RNA expansion segment 7L and assembly factor Noc2p specializes the ribosome biogenesis pathway between Saccharomyces cerevisiae and Candida albicans.Nucleic Acids Res. 2021 May 7;49(8):4655-4667. doi: 10.1093/nar/gkab218. Nucleic Acids Res. 2021. PMID: 33823547 Free PMC article.
-
Transcription-dependent recombination and the role of fork collision in yeast rDNA.Genes Dev. 2003 Jun 15;17(12):1497-506. doi: 10.1101/gad.1085403. Epub 2003 Jun 3. Genes Dev. 2003. PMID: 12783853 Free PMC article.
-
Ribosomal RNA gene repeats associate with the nuclear pore complex for maintenance after DNA damage.PLoS Genet. 2019 Apr 18;15(4):e1008103. doi: 10.1371/journal.pgen.1008103. eCollection 2019 Apr. PLoS Genet. 2019. PMID: 30998688 Free PMC article.
-
Ribosomal DNA replication time coordinates completion of genome replication and anaphase in yeast.Cell Rep. 2023 Mar 28;42(3):112161. doi: 10.1016/j.celrep.2023.112161. Epub 2023 Feb 25. Cell Rep. 2023. PMID: 36842087 Free PMC article.
References
-
- Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. - PubMed
-
- Brewer B J, Fangman W L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. - PubMed
-
- Brewer B J, Lockshon D, Fangman W L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. - PubMed
-
- Defossez P A, Prusty R, Kaeberlein M, Lin S J, Ferrigno P, Silver P A, Keil R L, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
