Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae
- PMID: 11113194
- PMCID: PMC88793
- DOI: 10.1128/MCB.21.1.189-195.2001
Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae
Abstract
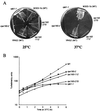
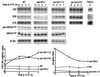
Temperature-sensitive RNA polymerase III (rpc160-112 and rpc160-270) mutants were analyzed for the synthesis of tRNAs and rRNAs in vivo, using a double-isotopic-labeling technique in which cells are pulse-labeled with [(33)P]orthophosphate and coextracted with [(3)H]uracil-labeled wild-type cells. Individual RNA species were monitored by Northern blot hybridization or amplified by reverse transcription. These mutants impaired the synthesis of RNA polymerase III transcripts with little or no influence on mRNA synthesis but also largely turned off the formation of the 25S, 18S, and 5.8S mature rRNA species derived from the common 35S transcript produced by RNA polymerase I. In the rpc160-270 mutant, this parallel inhibition of tRNA and rRNA synthesis also occurred at the permissive temperature (25 degrees C) and correlated with an accumulation of 20S pre-rRNA. In the rpc160-112 mutant, inhibition of rRNA synthesis and the accumulation of 20S pre-rRNA were found only at 37 degrees C. The steady-state rRNA/tRNA ratio of these mutants reflected their tRNA and rRNA synthesis pattern: the rpc160-112 mutant had the threefold shortage in tRNA expected from its preferential defect in tRNA synthesis at 25 degrees C, whereas rpc160-270 cells completely adjusted their rRNA/tRNA ratio down to a wild-type level, consistent with the tight coupling of tRNA and rRNA synthesis in vivo. Finally, an RNA polymerase I (rpa190-2) mutant grown at the permissive temperature had an enhanced level of pre-tRNA, suggesting the existence of a physiological coupling between rRNA synthesis and pre-tRNA processing.
Figures



References
-
- Bredow S, Kleinert H, Benecke B J. Sequence and factor requirements for faithful in vitro transcription of human 7SL DNA. Gene. 1990;86:217–225. - PubMed
-
- Carles C, Riva M. Yeast RNA polymerase I subunits and genes. In: Paule M R, editor. Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. Berlin, Germany: Springer Verlag; 1998. pp. 9–38.
-
- Chédin S, Ferri M L, Peyroche G, Andrau J C, Jourdain S, Lefebvre O, Werner M, Carles C, Sentenac A. The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harbor Symp Quant Biol. 1998;63:381–389. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
