Position effects are influenced by the orientation of a transgene with respect to flanking chromatin
- PMID: 11113204
- PMCID: PMC88803
- DOI: 10.1128/MCB.21.1.298-309.2001
Position effects are influenced by the orientation of a transgene with respect to flanking chromatin
Abstract
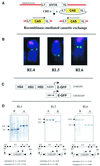
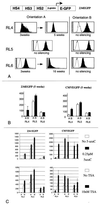
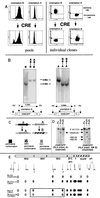
We have inserted two expression cassettes at tagged reference chromosomal sites by using recombinase-mediated cassette exchange in mammalian cells. The three sites of integration displayed either stable or silencing position effects that were dominant over the different enhancers present in the cassettes. These position effects were strongly dependent on the orientation of the construct within the locus, with one orientation being permissive for expression and the other being nonpermissive. Orientation-specific silencing, which was observed at two of the three site tested, was associated with hypermethylation but not with changes in chromatin structure, as judged by DNase I hypersensitivity assays. Using CRE recombinase, we were able to switch in vivo the orientation of the transgenes from the permissive to the nonpermissive orientation and vice versa. Switching from the permissive to the nonpermissive orientation led to silencing, but switching from the nonpermissive to the permissive orientation did not lead to reactivation of the transgene. Instead, transgene expression occurred dynamically by transcriptional oscillations, with 10 to 20% of the cells expressing at any given time. This result suggested that the cassette had been imprinted (epigenetically tagged) while it was in the nonpermissive orientation. Methylation analysis revealed that the methylation state of the inverted cassettes resembled that of silenced cassettes except that the enhancer had selectively lost some of its methylation. Sorting of the expressing and nonexpressing cell populations provided evidence that the transcriptional oscillations of the epigenetically tagged cassette are associated with changes in the methylation status of regulatory elements in the transgene. This suggests that transgene methylation is more dynamic than was previously assumed.
Figures


Similar articles
-
The human beta-globin locus control region can silence as well as activate gene expression.Mol Cell Biol. 2005 May;25(10):3864-74. doi: 10.1128/MCB.25.10.3864-3874.2005. Mol Cell Biol. 2005. PMID: 15870261 Free PMC article.
-
Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange.J Mol Biol. 1999 Oct 1;292(4):779-85. doi: 10.1006/jmbi.1999.3113. J Mol Biol. 1999. PMID: 10525404
-
Functional dissection of the mouse tyrosinase locus control region identifies a new putative boundary activity.Nucleic Acids Res. 2003 Nov 1;31(21):6290-305. doi: 10.1093/nar/gkg793. Nucleic Acids Res. 2003. PMID: 14576318 Free PMC article.
-
Ubiquitous Chromatin-opening Elements (UCOEs): Applications in biomanufacturing and gene therapy.Biotechnol Adv. 2017 Sep;35(5):557-564. doi: 10.1016/j.biotechadv.2017.05.004. Epub 2017 May 17. Biotechnol Adv. 2017. PMID: 28528197 Review.
-
Transcriptional transgene silencing and chromatin components.Plant Mol Biol. 2000 Jun;43(2-3):221-34. doi: 10.1023/a:1006483428789. Plant Mol Biol. 2000. PMID: 10999406 Review.
Cited by
-
The human beta-globin locus control region can silence as well as activate gene expression.Mol Cell Biol. 2005 May;25(10):3864-74. doi: 10.1128/MCB.25.10.3864-3874.2005. Mol Cell Biol. 2005. PMID: 15870261 Free PMC article.
-
Artificial chromosome-based transgenes in the study of genome function.Mamm Genome. 2006 Aug;17(8):791-807. doi: 10.1007/s00335-006-0023-9. Epub 2006 Aug 4. Mamm Genome. 2006. PMID: 16897340 Review.
-
DNA methylation density influences the stability of an epigenetic imprint and Dnmt3a/b-independent de novo methylation.Mol Cell Biol. 2002 Nov;22(21):7572-80. doi: 10.1128/MCB.22.21.7572-7580.2002. Mol Cell Biol. 2002. PMID: 12370304 Free PMC article.
-
hVGAT-mCherry: A novel molecular tool for analysis of GABAergic neurons derived from human pluripotent stem cells.Mol Cell Neurosci. 2015 Sep;68:244-57. doi: 10.1016/j.mcn.2015.08.007. Epub 2015 Aug 16. Mol Cell Neurosci. 2015. PMID: 26284979 Free PMC article.
-
Novel Strategies for Genomic Manipulation of Trichoderma reesei with the Purpose of Strain Engineering.Appl Environ Microbiol. 2015 Sep;81(18):6314-23. doi: 10.1128/AEM.01545-15. Epub 2015 Jul 6. Appl Environ Microbiol. 2015. PMID: 26150462 Free PMC article.
References
-
- Abremski K, Frommer B, Hoess R H. Linking-number changes in the DNA substrate during Cre-mediated loxP site-specific recombination. J Mol Biol. 1986;192:17–26. - PubMed
-
- Alami R, Greally J M, Tanimoto K, Hwang S, Feng Y Q, Engel J D, Fiering S, Bouhassira E E. Beta-globin YAC transgenes exhibit uniform expression levels but position effect variegation in mice. Hum Mol Genet. 2000;9:631–636. - PubMed
-
- Bergman Y, Mostoslavsky R. DNA demethylation: turning genes on. Biol Chem. 1998;379:401–407. - PubMed
-
- Bouhassira E E, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
