Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system
- PMID: 11114906
- PMCID: PMC94855
- DOI: 10.1128/JB.183.1.101-108.2001
Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system
Erratum in
- J Bacteriol 2001 May;183(9):
Abstract
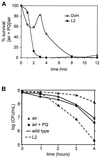
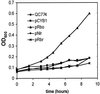
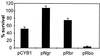
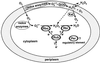
Evidence is presented for an alternative to the superoxide dismutase (SOD)-catalase oxidative stress defense system in Desulfovibrio vulgaris (strain Hildenborough). This alternative system consists of the nonheme iron proteins, rubrerythrin (Rbr) and rubredoxin oxidoreductase (Rbo), the product of the rbo gene (also called desulfoferrodoxin). A Deltarbo strain of D. vulgaris was found to be more sensitive to internal superoxide exposure than was the wild type. Unlike Rbo, expression of plasmid-borne Rbr failed to restore the aerobic growth of a SOD-deficient strain of Escherichia coli. Conversely, plasmid-borne expression of two different Rbrs from D. vulgaris increased the viability of a catalase-deficient strain of E. coli that had been exposed to hydrogen peroxide whereas Rbo actually decreased the viability. A previously undescribed D. vulgaris gene was found to encode a protein having 50% sequence identity to that of E. coli Fe-SOD. This gene also encoded an extended N-terminal sequence with high homologies to export signal peptides of periplasmic redox proteins. The SOD activity of D. vulgaris is not affected by the absence of Rbo and is concentrated in the periplasmic fraction of cell extracts. These results are consistent with a superoxide reductase rather than SOD activity of Rbo and with a peroxidase activity of Rbr. A joint role for Rbo and Rbr as a novel cytoplasmic oxidative stress protection system in D. vulgaris and other anaerobic microorganisms is proposed.
Figures



References
-
- Alban P S, Krieg N R. A hydrogen peroxide resistant mutant of Spirillum volutans has NADH peroxidase activity but no increased oxygen tolerance. Can J Microbiol. 1998;44:87–91.
-
- Alban P S, Popham D L, Rippere K E, Krieg N R. Identification of a gene for a rubrerythrin/nigerythrin-like protein from Sprillum volutans by using amino acid sequence data from mass spectrometry and NH2-terminal sequencing. J Appl Microbiol. 1998;85:875–882. - PubMed
-
- Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Green Publishing and Wiley-Interscience; 1990.
-
- Barnes A C, Balebona M C, Horne M T, Ellis A E. Superoxide dismutase and catalase in Photobacterium damselae subsp. piscicida and their roles in resistance to reactive oxygen species. Microbiology. 1999;145:483–494. - PubMed
-
- Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

