Characterization and evolution of anthranilate 1,2-dioxygenase from Acinetobacter sp. strain ADP1
- PMID: 11114907
- PMCID: PMC94856
- DOI: 10.1128/JB.183-1.109-118.2001
Characterization and evolution of anthranilate 1,2-dioxygenase from Acinetobacter sp. strain ADP1
Abstract
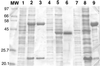
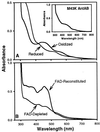
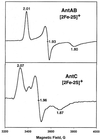
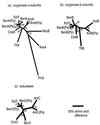
The two-component anthranilate 1,2-dioxygenase of the bacterium Acinetobacter sp. strain ADP1 was expressed in Escherichia coli and purified to homogeneity. This enzyme converts anthranilate (2-aminobenzoate) to catechol with insertion of both atoms of O(2) and consumption of one NADH. The terminal oxygenase component formed an alpha(3)beta(3) hexamer of 54- and 19-kDa subunits. Biochemical analyses demonstrated one Rieske-type [2Fe-2S] center and one mononuclear nonheme iron center in each large oxygenase subunit. The reductase component, which transfers electrons from NADH to the oxygenase component, was found to contain approximately one flavin adenine dinucleotide and one ferredoxin-type [2Fe-2S] center per 39-kDa monomer. Activities of the combined components were measured as rates and quantities of NADH oxidation, substrate disappearance, product appearance, and O(2) consumption. Anthranilate conversion to catechol was stoichiometrically coupled to NADH oxidation and O(2) consumption. The substrate analog benzoate was converted to a nonaromatic benzoate 1,2-diol with similarly tight coupling. This latter activity is identical to that of the related benzoate 1, 2-dioxygenase. A variant anthranilate 1,2-dioxygenase, previously found to convey temperature sensitivity in vivo because of a methionine-to-lysine change in the large oxygenase subunit, was purified and characterized. The purified M43K variant, however, did not hydroxylate anthranilate or benzoate at either the permissive (23 degrees C) or nonpermissive (39 degrees C) growth temperatures. The wild-type anthranilate 1,2-dioxygenase did not efficiently hydroxylate methylated or halogenated benzoates, despite its sequence similarity to broad-substrate specific dioxygenases that do. Phylogenetic trees of the alpha and beta subunits of these terminal dioxygenases that act on natural and xenobiotic substrates indicated that the subunits of each terminal oxygenase evolved from a common ancestral two-subunit component.
Figures


References
-
- Altenschmidt U, Fuchs G. Novel aerobic 2-aminobenzoate metabolism. Eur J Biochem. 1992;205:721–727. - PubMed
-
- Assinder S J, Williams P A. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. - PubMed
-
- Batie C J, LaHaie E, Ballou D P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J Biol Chem. 1987;262:1510–1518. - PubMed
-
- Bertini I, Cremonini M A, Ferretti S, Lozzi I, Luchionat C, Viezzoli M S. Arene hydroxylases: metalloenzymes catalysing dioxygenation of aromatic compounds. Coord Chem Rev. 1996;151:145–160.
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

