The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors
- PMID: 11115874
- PMCID: PMC59855
- DOI: 10.1104/pp.124.4.1558
The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors
Abstract
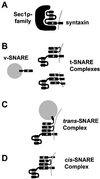
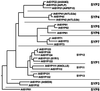
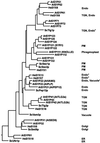
Many factors have been characterized as essential for vesicle trafficking, including a number of proteins commonly referred to as soluble N-ethylmaleimide-sensitive factor adaptor protein receptor (SNARE) components. The Arabidopsis genome contains a remarkable number of SNAREs. In general, the vesicle fusion machinery appears highly conserved. However, whereas some classes of yeast and mammalian genes appear to be lacking in Arabidopsis, this small plant genome has gene families not found in other eukaryotes. Very little is known about the precise function of plant SNAREs. By contrast, the intracellular localization of and interactions between a large number of plant SNAREs have been determined, and these data are discussed in light of the phylogenetic analysis.
Figures


References
-
- Aalto MK, Ruohonen L, Hosono K, Keranen S. Cloning and sequencing of the yeast Saccharomyces cerevisiae SEC1 gene localized on chromosome IV. Yeast. 1991;7:643–650. - PubMed
-
- Abeliovich H, Grote E, Novick P, Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J Biol Chem. 1998;273:11719–11727. - PubMed
-
- Adams MD. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

