Characterization of the response of the Arabidopsis response regulator gene family to cytokinin
- PMID: 11115887
- PMCID: PMC59868
- DOI: 10.1104/pp.124.4.1706
Characterization of the response of the Arabidopsis response regulator gene family to cytokinin
Abstract
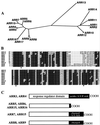
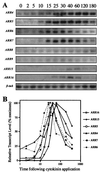
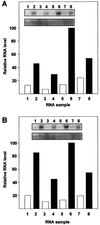
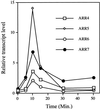
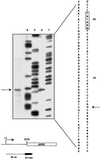
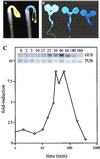
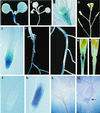
We examined the expression of a family of Arabidopsis response regulators (ARR) and found that the steady-state levels of RNA for most are elevated very rapidly by cytokinin. Using nuclear run-on assays we demonstrated that this increase in ARR transcript levels in response to cytokinin is due, at least in part, to increased transcription. The start site of transcription for the ARR5 gene was identified using primer extension analysis. A DNA fragment comprised of 1.6 kb upstream of the ARR5 transcript start site conferred cytokinin-inducible gene expression when fused to a beta-glucuronidase reporter, confirming that the transcription rate of ARR5 is elevated by cytokinin. This reporter construct was also used to examine the spatial pattern of ARR5 expression. The highest levels of expression were observed in the root and shoot apical meristems, at the junction of the pedicle and the silique, and in the central portion of mature roots. The expression of ARR5 in the apical meristems was confirmed by whole mount in situ analysis of seedlings and is consistent with a role for cytokinin in regulating cell division in vivo.
Figures
Similar articles
-
Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity.Plant Cell. 2003 Nov;15(11):2532-50. doi: 10.1105/tpc.014928. Epub 2003 Oct 10. Plant Cell. 2003. PMID: 14555694 Free PMC article.
-
Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis.Plant J. 2000 Dec;24(5):559-67. doi: 10.1046/j.1365-313x.2000.00893.x. Plant J. 2000. PMID: 11123795
-
Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses.Plant J. 2004 Apr;38(2):203-14. doi: 10.1111/j.1365-313X.2004.02038.x. Plant J. 2004. PMID: 15078325
-
Isolation and characterization of the orchid cytokinin oxidase DSCKX1 promoter.J Exp Bot. 2002 Sep;53(376):1899-907. doi: 10.1093/jxb/erf055. J Exp Bot. 2002. PMID: 12177129
-
Expression pattern of diacylglycerol acyltransferase-1, an enzyme involved in triacylglycerol biosynthesis, in Arabidopsis thaliana.Plant Mol Biol. 2003 May;52(1):31-41. doi: 10.1023/a:1023935605864. Plant Mol Biol. 2003. PMID: 12825687
Cited by
-
Crosstalk of Cytokinin with Ethylene and Auxin for Cell Elongation Inhibition and Boron Transport in Arabidopsis Primary Root under Boron Deficiency.Plants (Basel). 2022 Sep 8;11(18):2344. doi: 10.3390/plants11182344. Plants (Basel). 2022. PMID: 36145745 Free PMC article.
-
Tissue- and Cell-Specific Cytokinin Activity in Populus × canescens Monitored by ARR5::GUS Reporter Lines in Summer and Winter.Front Plant Sci. 2016 May 13;7:652. doi: 10.3389/fpls.2016.00652. eCollection 2016. Front Plant Sci. 2016. PMID: 27242853 Free PMC article.
-
Genome-wide identification and expression analysis of the TaRRA gene family in wheat (Triticum aestivum L.).Front Plant Sci. 2022 Aug 30;13:1006409. doi: 10.3389/fpls.2022.1006409. eCollection 2022. Front Plant Sci. 2022. PMID: 36110359 Free PMC article.
-
Programmed cell death induced by high levels of cytokinin in Arabidopsis cultured cells is mediated by the cytokinin receptor CRE1/AHK4.J Exp Bot. 2012 Apr;63(7):2825-32. doi: 10.1093/jxb/ers008. Epub 2012 Feb 6. J Exp Bot. 2012. PMID: 22312114 Free PMC article.
-
Identification and expression of cytokinin signaling and meristem identity genes in sulfur deficient grapevine (Vitis vinifera L.).Plant Signal Behav. 2009 Dec;4(12):1128-35. doi: 10.4161/psb.4.12.9942. Plant Signal Behav. 2009. PMID: 20514227 Free PMC article.
References
-
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994.
-
- Chaconas G, van de Sande JH. 5′ 32P labeling of RNA and DNA restriction fragments. Methods Enzymol. 1980;65:75–88. - PubMed
-
- Chang C, Kwok SF, Bleecker A, Meyerowitz E. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

