Aux/IAA proteins are phosphorylated by phytochrome in vitro
- PMID: 11115889
- PMCID: PMC59870
- DOI: 10.1104/pp.124.4.1728
Aux/IAA proteins are phosphorylated by phytochrome in vitro
Abstract
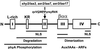
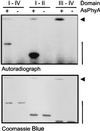
Auxin/indole-3-acetic acid (Aux/IAA) genes encode short-lived transcription factors that are induced as a primary response to the plant growth hormone IAA or auxin. Gain-of-function mutations in Arabidopsis genes, SHY2/IAA3, AXR3/IAA17, and AXR2/IAA7 cause pleiotropic phenotypes consistent with enhanced auxin responses, possibly by increasing Aux/IAA protein stability. Semidominant mutations shy2-1D, shy2-2, axr3-1, and axr2-1 induce ectopic light responses in dark-grown seedlings. Because genetic studies suggest that the shy2-1D and shy2-2 mutations bypass phytochrome requirement for certain aspects of photomorphogenesis, we tested whether SHY2/IAA3 and related Aux/IAA proteins interact directly with phytochrome and whether they are substrates for its protein kinase activity. Here we show that recombinant Aux/IAA proteins from Arabidopsis and pea (Pisum sativum) interact in vitro with recombinant phytochrome A from oat (Avena sativa). We further show that recombinant SHY2/IAA3, AXR3/IAA17, IAA1, IAA9, and Ps-IAA4 are phosphorylated by recombinant oat phytochrome A in vitro. Deletion analysis of Ps-IAA4 indicates that phytochrome A phosphorylation occurs on the N-terminal half of the protein. Metabolic labeling and immunoprecipitation studies with affinity-purified antibodies to IAA3 demonstrate increased in vivo steady-state levels of mutant IAA3 in shy2-2 plants and phosphorylation of the SHY2-2 protein in vivo. Phytochrome-dependent phosphorylation of Aux/IAA proteins is proposed to provide one molecular mechanism for integrating auxin and light signaling in plant development.
Figures





References
-
- Abel S, Nguyen D, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. - PubMed
-
- Abel S, Theologis A. A polymorphic bipartite motif signals nuclear targeting of early auxin-induced proteins related to PS-IAA4 from pea (Pisum sativum) Plant J. 1995;8:87–96. - PubMed
-
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1:939–948. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

