Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases
- PMID: 11115898
- PMCID: PMC59879
- DOI: 10.1104/pp.124.4.1844
Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases
Abstract
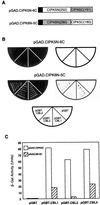
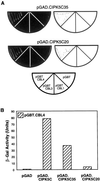
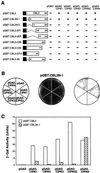
Calcium is a critical component in a number of plant signal transduction pathways. A new family of calcium sensors called calcineurin B-like proteins (AtCBLs) have been recently identified from Arabidopsis. These calcium sensors have been shown to interact with a family of protein kinases (CIPKs). Here we report that each individual member of AtCBL family specifically interacts with a subset of CIPKs and present structural basis for the interaction and for the specificity underlying these interactions. Although the C-terminal region of CIPKs is responsible for interaction with AtCBLs, the N-terminal region of CIPKs is also involved in determining the specificity of such interaction. We have also shown that all three EF-hand motifs in AtCBL members are required for the interaction with CIPKs. Several AtCBL members failed to interact with any of the CIPKs presented in this study, suggesting that these AtCBL members either have other CIPKs as targets or they target distinct proteins other than CIPKs. These results may provide structural basis for the functional specificity of CBL family of calcium sensors and their targets.
Figures


References
-
- Ames JB, Ishima R, Tanaka T, Gordon JI, Stryer L, Ikura M. Molecular mechanics of calcium-myristoyl switches. Nature. 1997;389:198–202. - PubMed
-
- Braunewell K-H, Gundelfinger ED. Intracellular neuronal calcium sensor proteins: a family of EF-hand calcium binding proteins in search of a function. Cell Tissue Res. 1999;295:1–12. - PubMed
-
- Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. - PubMed
-
- Bush DS. Calcium regulation in plant cells and its role in signaling. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:95–122.
-
- Calvert PD, Klenchin VA, Bownds MD. Rhodopsin kinase inhibition by recoverin: function of recoverin myristoylation. J Biol Chem. 1995;270:24127–24129. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

