A random sequencing approach for the analysis of the Trypanosoma cruzi genome: general structure, large gene and repetitive DNA families, and gene discovery
- PMID: 11116094
- PMCID: PMC313047
- DOI: 10.1101/gr.gr-1463r
A random sequencing approach for the analysis of the Trypanosoma cruzi genome: general structure, large gene and repetitive DNA families, and gene discovery
Abstract
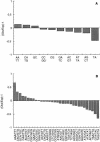
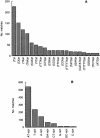
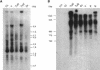
A random sequence survey of the genome of Trypanosoma cruzi, the agent of Chagas disease, was performed and 11,459 genomic sequences were obtained, resulting in approximately 4.3 Mb of readable sequences or approximately 10% of the parasite haploid genome. The estimated total GC content was 50.9%, with a high representation of A and T di- and trinucleotide repeats. Out of the estimated 5000 parasite genes, 947 putative new genes were identified. Another 1723 sequences corresponded to genes detected previously in T. cruzi through expression sequence tag analysis. 7735 sequences had no matches in the database, but the presence of open reading frames that passed Fickett's test suggests that some might contain coding DNA. The survey was highly redundant, with approximately 35% of the sequences included in a few large sequence families. Some of them code for protein families present in dozens of copies, including proteins essential for parasite survival and retrotransposons. Other sequence families include repetitive DNA present in thousands of copies per haploid genome. Some families in the latter group are new, parasite-specific, repetitive DNAs. These results suggest that T. cruzi could constitute an interesting model to analyze gene and genome evolution due to its plasticity in terms of sequence amplification and divergence. Additional information can be found at http://www.iib.unsam.edu.ar/tcruzi.gss. html.
Figures

References
-
- Araya J, Cano MI, Gomes HB, Novak EM, Requena JM, Alonso C, Levin MJ, Guevara P, Ramirez JL, Da Silveira JF. Characterization of an interspersed repetitive DNA element in the genome of Trypanosoma cruzi. Parasitology. 1997;115:563–570. - PubMed
-
- Armah DA, Mensa-Wilmot K. S-myristoylation of a glycosylphosphatidylinositol-specific phospholipase C in Trypanosoma brucei. J Biol Chem. 1999;274:5931–5938. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
