P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL
- PMID: 11118214
- PMCID: PMC305888
- DOI: 10.1093/emboj/19.24.6792
P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL
Abstract
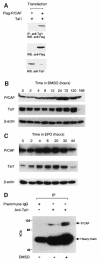
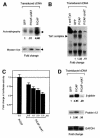
The basic helix-loop-helix transcription factor TAL1 (or SCL) is a critical regulator of hematopoietic and vascular development and is misexpressed in the majority of patients with T-cell acute lymphoblastic leukemia. We found previously that TAL1 could interact with transcriptional co-activator and co-repressor complexes possessing histone acetyltransferase and deacetylase activities, respectively. Here, we report that TAL1 is subject to acetylation in vivo and can be acetylated by p300 and the p300/CBP-associated factor P/CAF in vitro. P/CAF-mediated acetylation, which mapped to a lysine-rich motif in the loop region, increased TAL1 binding to DNA while selectively inhibiting its interaction with the transcriptional co-repressor mSin3A. Furthermore, P/CAF protein, TAL1-P/CAF interaction and TAL1 acetylation increased significantly in murine erythroleukemia cells induced to differentiate in culture, while enforced expression of an acetylation-defective P/CAF mutant inhibited endogenous TAL1 acetylation, TAL1 DNA-binding activity, TAL1-directed transcription and terminal differentiation of these cells. These results reveal a novel mechanism by which TAL1 activity is regulated and implicate acetylation of this transcription factor in promotion of erythroid differentiation.
Figures




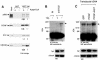
Similar articles
-
Role of helix-loop-helix proteins during differentiation of erythroid cells.Mol Cell Biol. 2011 Apr;31(7):1332-43. doi: 10.1128/MCB.01186-10. Epub 2011 Jan 31. Mol Cell Biol. 2011. PMID: 21282467 Free PMC article. Review.
-
mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor.Mol Cell Biol. 2000 Mar;20(6):2248-59. doi: 10.1128/MCB.20.6.2248-2259.2000. Mol Cell Biol. 2000. PMID: 10688671 Free PMC article.
-
p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein.Oncogene. 1999 Sep 2;18(35):4958-67. doi: 10.1038/sj.onc.1202889. Oncogene. 1999. PMID: 10490830
-
Interaction of EVI1 with cAMP-responsive element-binding protein-binding protein (CBP) and p300/CBP-associated factor (P/CAF) results in reversible acetylation of EVI1 and in co-localization in nuclear speckles.J Biol Chem. 2001 Nov 30;276(48):44936-43. doi: 10.1074/jbc.M106733200. Epub 2001 Sep 21. J Biol Chem. 2001. PMID: 11568182
-
CBP and p300: versatile coregulators with important roles in hematopoietic gene expression.J Leukoc Biol. 2002 Apr;71(4):545-56. J Leukoc Biol. 2002. PMID: 11927640 Review. No abstract available.
Cited by
-
Role of helix-loop-helix proteins during differentiation of erythroid cells.Mol Cell Biol. 2011 Apr;31(7):1332-43. doi: 10.1128/MCB.01186-10. Epub 2011 Jan 31. Mol Cell Biol. 2011. PMID: 21282467 Free PMC article. Review.
-
Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic leukemia zinc finger protein.Mol Cell Biol. 2005 Jul;25(13):5552-66. doi: 10.1128/MCB.25.13.5552-5566.2005. Mol Cell Biol. 2005. PMID: 15964811 Free PMC article.
-
LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis.Proc Natl Acad Sci U S A. 2009 Jun 23;106(25):10141-6. doi: 10.1073/pnas.0900437106. Epub 2009 Jun 3. Proc Natl Acad Sci U S A. 2009. PMID: 19497860 Free PMC article.
-
Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications.Genes Dev. 2005 Aug 15;19(16):1885-93. doi: 10.1101/gad.1333905. Genes Dev. 2005. PMID: 16103216 Free PMC article.
-
Targets of the Tal1 transcription factor in erythrocytes: E2 ubiquitin conjugase regulation by Tal1.J Biol Chem. 2010 Feb 19;285(8):5338-46. doi: 10.1074/jbc.M109.030296. Epub 2009 Dec 22. J Biol Chem. 2010. PMID: 20028976 Free PMC article.
References
-
- Akazawa C., Ishibashi,M., Shimizu,C., Nakanishi,S. and Kageyama,R. (1995) A mammalian helix–loop–helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem., 270, 8730–8738. - PubMed
-
- Arany Z., Newsome,D., Oldread,E., Livingston,D.M. and Eckner,R. (1995) A family of transcriptional adaptor proteins targeted by the E1A protein. Nature, 374, 81–84. - PubMed
-
- Atchley W.R., Wollenberg,K.R., Fitch,W.M., Terhalle,W. and Dress,A.W. (2000) Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol. Biol. Evol., 17, 164–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

