Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase
- PMID: 11118450
- PMCID: PMC2606050
- DOI: 10.1074/jbc.M009901200
Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase
Abstract
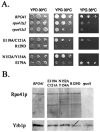
Alignment of three fungal mtRNA polymerases revealed conserved amino acid sequences in an amino-terminal region of the Saccharomyces cerevisiae enzyme implicated previously as harboring an important functional domain. Phenotypic analysis of deletion and point mutations, in conjunction with a yeast two-hybrid assay, revealed that Nam1p, a protein involved in RNA processing and translation in mitochondria, binds specifically to this domain. The significance of this interaction in vivo was demonstrated by the fact that the temperature-sensitive phenotype of a deletion mutation (rpo41Delta2), which impinges on this amino-terminal domain, is suppressed by overproducing Nam1p. In addition, mutations in the amino-terminal domain result specifically in decreased steady-state levels of mature mitochondrial CYTB and COXI transcripts, which is a primary defect observed in NAM1 null mutant yeast strains. Finally, one point mutation (R129D) did not abolish Nam1p binding, yet displayed an obvious COX1/CYTB transcript defect. This mutation exhibited the most severe mitochondrial phenotype, suggesting that mutations in the amino-terminal domain can perturb other critical interactions, in addition to Nam1p binding, that contribute to the observed phenotypes. These results implicate the amino-terminal domain of mtRNA polymerases in coupling additional factors and activities involved in mitochondrial gene expression directly to the transcription machinery.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

