Legionella pneumophila major acid phosphatase and its role in intracellular infection
- PMID: 11119504
- PMCID: PMC97870
- DOI: 10.1128/IAI.69.1.177-185.2001
Legionella pneumophila major acid phosphatase and its role in intracellular infection
Abstract
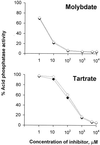
Legionella pneumophila is an intracellular pathogen of protozoa and alveolar macrophages. This bacterium contains a gene (pilD) that is involved in both type IV pilus biogenesis and type II protein secretion. We previously demonstrated that the PilD prepilin peptidase is crucial for intracellular infection by L. pneumophila and that the secreted pilD-dependent proteins include a metalloprotease, an acid phosphatase, an esterase/lipase, a phospholipase A, and a p-nitrophenyl phosphorylcholine hydrolase. Since mutants lacking type IV pili, the protease, or the phosphorylcholine hydrolase are not defective for intracellular infection, we sought to determine the significance of the secreted acid phosphatase activity. Three mutants defective in acid phosphatase activity were isolated from a population of mini-Tn10-mutagenized L. pneumophila. Supernatants as well as cell lysates from these mutants contained minimal acid phosphatase activity while possessing normal levels of other pilD-dependent exoproteins. Genetic studies indicated that the gene affected by the transposon insertions encoded a novel bacterial histidine acid phosphatase, which we designated Map for major acid phosphatase. Subsequent inhibitor studies indicated that Map, like its eukaryotic homologs, is a tartrate-sensitive acid phosphatase. The map mutants grew within macrophage-like U937 cells and Hartmannella amoebae to the same degree as did wild-type legionellae, indicating that this acid phosphatase is not essential for L. pneumophila intracellular infection. However, in the course of characterizing our new mutants, we gained evidence for a second pilD-dependent acid phosphatase activity that, unlike Map, is tartrate resistant.
Figures







References
-
- Abu Kwaik Y. Fatal attraction of mammalian cells to Legionella pneumophila. Mol Microbiol. 1998;30:689–695. - PubMed
-
- Alm R A, Hallinan J P, Watson A A, Mattick J S. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

