Dominance of virus over host factors in cross-species activation of human cytomegalovirus early gene expression
- PMID: 11119570
- PMCID: PMC113894
- DOI: 10.1128/JVI.75.1.26-35.2001
Dominance of virus over host factors in cross-species activation of human cytomegalovirus early gene expression
Abstract
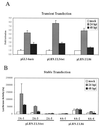
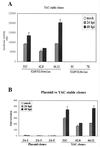
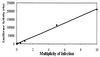
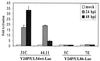
Human cytomegalovirus (HCMV) exhibits a highly restricted host range. In this study, we sought to examine the relative significance of host and viral factors in activating early gene expression of the HCMV UL54 (DNA polymerase) promoter in murine cells. Appropriate activation of the UL54 promoter at early times is essential for viral DNA replication. To study how the HCMV UL54 promoter is activated in murine cells, a transgenesis system based on yeast artificial chromosomes (YACs) was established for HCMV. A 178-kb YAC, containing a subgenomic fragment of HCMV encompassing the majority of the unique long (UL) region, was constructed by homologous recombination in yeast. This HCMV YAC backbone is defective for viral growth and lacks the major immediate-early (IE) gene region, thus permitting the analysis of essential cis-acting sequences when complemented in trans. To quantitatively measure the level of gene expression, we generated HCMV YACs containing a luciferase reporter gene inserted downstream of either the UL54 promoter or, as a control for late gene expression, the UL86 promoter, which directs expression of the major capsid protein. To determine the early gene activation pathway, point mutations were introduced into the inverted repeat 1 (IR1) element of the UL54 promoter of the HCMV YAC. In the transgenesis experiments, HCMV YACs and derivatives generated in yeast were introduced into NIH 3T3 murine cells by polyethylene glycol-mediated fusion. We found that infection of YAC, but not plasmid, transgenic lines with HCMV was sufficient to fully recapitulate the UL54 expression program at early times of infection, indicating the importance of remote regulatory elements in influencing regulation of the UL54 promoter. Moreover, YACs containing a mutant IR1 in the UL54 promoter led to reduced ( approximately 30-fold) reporter gene expression levels, indicating that HCMV major IE gene activation of the UL54 promoter is fully permissive in murine cells. In comparison with HCMV, infection of YAC transgenic NIH 3T3 lines with murine cytomegalovirus (MCMV) resulted in lower (more than one order of magnitude) efficiency in activating UL54 early gene expression. MCMV is therefore not able to fully activate HCMV early gene expression, indicating the significance of virus over host determinants in the cross-species activation of key early gene promoters. Finally, these studies show that YAC transgenesis can be a useful tool in functional analysis of viral proteins and control of gene expression for large viral genomes.
Figures





References
-
- Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan J S, Bittner A, Frueh K, Jackson M R, Peterson P A, Erlander M G, Ghazal P. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73:5757–5766. - PMC - PubMed
-
- DeMarchi J M. Nature of the block in the expression of some early virus genes in cells abortively infected with human cytomegalovirus. Virology. 1983;129:287–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

