The E1 initiator recognizes multiple overlapping sites in the papillomavirus origin of DNA replication
- PMID: 11119599
- PMCID: PMC113923
- DOI: 10.1128/JVI.75.1.292-302.2001
The E1 initiator recognizes multiple overlapping sites in the papillomavirus origin of DNA replication
Abstract
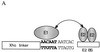
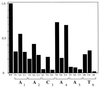
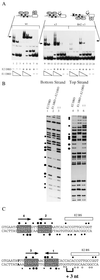
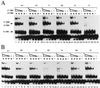
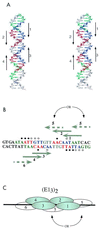
A common feature of replicator sequences from a variety of organisms is multiple binding sites for an initiator protein. By binding to the replicator, initiators mark the site and contribute to melting or distortion of the DNA. We have defined the recognition sequence for the papillomavirus E1 initiator and determined the arrangement of binding sites in the viral origin of replication. We show that E1 recognizes a hexanucleotide sequence which is present in overlapping arrays in virtually all papillomavirus replicators. Binding of the initiator to these sites would result in the formation of a closely packed array of E1 molecules that wrap around the double helix.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

