Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway
- PMID: 11119609
- PMCID: PMC113933
- DOI: 10.1128/JVI.75.1.408-419.2001
Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway
Abstract
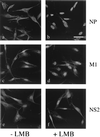
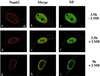
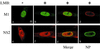
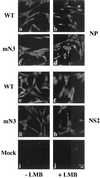
Influenza virus transcription occurs in the nuclei of infected cells, where the viral genomic RNAs are complexed with a nucleoprotein (NP) to form ribonucleoprotein (RNP) structures. Prior to assembly into progeny virions, these RNPs exit the nucleus and accumulate in the cytoplasm. The mechanisms responsible for RNP export are only partially understood but have been proposed to involve the viral M1 and NS2 polypeptides. We found that the drug leptomycin B (LMB), which specifically inactivates the cellular CRM1 polypeptide, caused nuclear retention of NP in virus-infected cells, indicating a role for the CRM1 nuclear export pathway in RNP egress. However, no alteration was seen in the cellular distribution of M1 or NS2, even in the case of a mutant virus which synthesizes greatly reduced amounts of NS2. Furthermore, NP was distributed throughout the nuclei of infected cells at early times postinfection but, when retained in the nucleus at late times by LMB treatment, was redistributed to the periphery of the nucleoplasm. No such change was seen in the nuclear distribution of M1 or NS2 after drug treatment. Similar to the behavior of NP, M1 and NS2 in infected cells, LMB treatment of cells expressing each polypeptide in isolation caused nuclear retention of NP but not M1 or NS2. Conversely, overexpression of CRM1 caused increased cytoplasmic accumulation of NP but had little effect on M1 or NS2 distribution. Consistent with this, NP bound CRM1 in vitro. Overall, these data raise the possibility that RNP export is mediated by a direct interaction between NP and the cellular CRM1 export pathway.
Figures



References
-
- Askjaer P, Heick Jensen T, Nilsson J, Engelmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. - PubMed
-
- Briedis D J, Conti G, Munn E A, Mahy B W. Migration of influenza virus-specific polypeptides from cytoplasm to nucleus of infected cells. Virology. 1981;111:154–164. - PubMed
-
- Bullido R, Gomez-Puertas P, Albo C, Portela A. Several protein regions contribute to determine the nuclear and cytoplasmic localization of the influenza A virus nucleopotein. J Gen Virol. 2000;81:135–142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

