T cell receptor (TCR)-mediated repertoire selection and loss of TCR vbeta diversity during the initiation of a CD4(+) T cell response in vivo
- PMID: 11120769
- PMCID: PMC2213496
- DOI: 10.1084/jem.192.12.1719
T cell receptor (TCR)-mediated repertoire selection and loss of TCR vbeta diversity during the initiation of a CD4(+) T cell response in vivo
Abstract
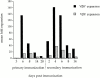
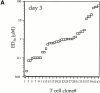
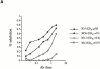
We recently described a novel way to isolate populations of antigen-reactive CD4(+) T cells with a wide range of reactivity to a specific antigen, using immunization with a fixed dose of nominal antigen and FACS((R)) sorting by CD4(high) expression. Phenotypic, FACS((R)), functional, antibody inhibition, and major histocompatibility complex-peptide tetramer analyses, as well as T cell receptor Vbeta sequence analyses, of the antigen-specific CD4(high) T cell populations demonstrated that a diverse sperm whale myoglobin 110-121-reactive CD4(+) T cell repertoire was activated at the beginning (day 3 after immunization) of the immune response. Within 6 d of immunization, lower affinity clones were lost from the responding population, leaving an expanded population of oligoclonal, intermediate affinity (and residual high affinity) T cells. This T cell subset persisted for at least 4 wk after immunization and dominated the secondary immune response. These data provide evidence that CD4(+) T cell repertoire selection occurs early in the immune response in vivo and suggest that persistence and expansion of a population of oligoclonal, intermediate affinity T cells is involved in CD4(+) T cell memory.
Figures







References
-
- Kim D.T., Rothbard J.B., Bloom D.D., Fathman C.G. Quantitative analysis of T cell activationrole of TCR/ligand density and TCR affinity. J. Immunol. 1996;156:2737–2742. - PubMed
-
- Sloan-Lancaster J., Allen P.M. Altered peptide ligand-induced partial T cell activationmolecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 1996;14:1–27. - PubMed
-
- Hampl J., Chien Y.H., Davis M.M. CD4 augments the response of a T cell to agonist but not to antagonist ligands. Immunity. 1997;7:379–385. - PubMed
-
- Kersh G.J., Allen P.M. Essential flexibility in the T-cell recognition of antigen. Nature. 1996;380:495–498. - PubMed
-
- Garcia K.C., Degano M., Pease L.R., Huang M., Peterson P.A., Teyton L., Wilson I.A. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 1998;279:1166–1172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

