A large upstream region is not necessary for gene expression or hypersensitive site formation at the mouse beta -globin locus
- PMID: 11121056
- PMCID: PMC18957
- DOI: 10.1073/pnas.97.26.14554
A large upstream region is not necessary for gene expression or hypersensitive site formation at the mouse beta -globin locus
Abstract
Developmental expression at the beta-globin locus is regulated in part by the locus control region, a region upstream of the genes containing at least five major DNase I hypersensitive sites (HSs) in mammalian erythrocytes. Sequences farther 5' of these HSs are conserved in mouse and human, and both loci are embedded within a cluster of functional odorant receptor genes. In humans, distant upstream sequences have been implicated in regulation of the beta-globin genes. In this study, the role of the 5'-most HSs and their adjacent sequence was investigated by deletion of an 11-kb region from the mouse locus, including 5'HS 4.2, 5'HS 5, 5'HS 6, and the 5'beta1 odorant receptor gene. Mice that were homozygous for this deletion were fully viable, and no significant effect on adult beta-globin gene expression was seen. 5'HSs 1-4, which are located downstream of the deletion, were still present in the mutant mice. In addition, two new upstream HSs, HS -60.7 and HS -62.5, were found in erythroid tissue of both wild-type and mutant mice. Therefore, although the possibility of a minor role still exists, neither the HSs nor the other regions deleted in this study are essential for beta-globin gene expression, and it is unlikely that chromatin structure is affected either upstream or downstream of the deletion. This is the largest deletion at the mouse locus control region to show no apparent phenotype, and focuses attention on the possible contribution of sequences even farther upstream.
Figures
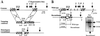
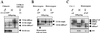



References
-
- Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F. Genes Dev. 1991;5:1387–1394. - PubMed
-
- Martin D I, Fiering S, Groudine M. Curr Opin Genet Dev. 1996;6:488–495. - PubMed
-
- Hardison R, Slightom J L, Gumucio D L, Goodman M, Stojanovic N, Miller W. Gene. 1997;205:73–94. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

