Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers
- PMID: 11121058
- PMCID: PMC18959
- DOI: 10.1073/pnas.97.26.14566
Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers
Abstract
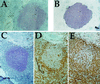
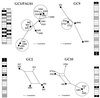
The tremendous dynamics of HIV infection finds expression in the tempo of sequence diversification. Genetic diversity calculations require the clearance of a majority of infected cells, the obvious predator being anti-HIV immune responses. Indeed, infiltration of germinal centers (GCs) by HIV-specific CD8(+) cytotoxic T lymphocytes has been described. A corollary to this description would be limited diffusion of virus within lymphoid structures. HIV efficiently infects and replicates mainly in activated CD4(+) T lymphoblasts. These cells are found within GCs after their activation in the adjacent periarteriolar lymphoid sheath (PALS). Here GCs and PALS have been dissected from consecutive 10-micrometer sections through splenic tissue from three HIV-1-infected patients. Nested PCR amplification of the two first hypervariable regions of the env gene indicated that 38-78% of sections contained HIV-infected cells. Since there are several hundred CD4(+) T cells per GC section, approximately 0.09-0.64% harbor proviral DNA. Such a low frequency not only suggests that virions on the follicular dendritic cell surfaces do not readily infect adjacent T cells but also indicates highly restricted spread of HIV within GCs and the PALS. Sections were heavily infiltrated by CD8(+) cells, which, together with a large body of extant data, suggests that the majority of infected cells are destroyed by HIV-specific cytotoxic T lymphocytes before becoming productively infected. Finally, sequence analysis revealed that those HIV-positive cells were multiply infected, which helps explain widespread recombination despite a low overall frequency of infected cells.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

