Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology
- PMID: 11121445
- PMCID: PMC2190600
- DOI: 10.1083/jcb.151.6.1321
Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology
Abstract
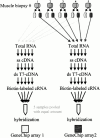
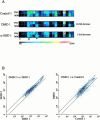
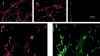
We used expression profiling to define the pathophysiological cascades involved in the progression of two muscular dystrophies with known primary biochemical defects, dystrophin deficiency (Duchenne muscular dystrophy) and alpha-sarcoglycan deficiency (a dystrophin-associated protein). We employed a novel protocol for expression profiling in human tissues using mixed samples of multiple patients and iterative comparisons of duplicate datasets. We found evidence for both incomplete differentiation of patient muscle, and for dedifferentiation of myofibers to alternative lineages with advancing age. One developmentally regulated gene characterized in detail, alpha-cardiac actin, showed abnormal persistent expression after birth in 60% of Duchenne dystrophy myofibers. The majority of myofibers ( approximately 80%) remained strongly positive for this protein throughout the course of the disease. Other developmentally regulated genes that showed widespread overexpression in these muscular dystrophies included embryonic myosin heavy chain, versican, acetylcholine receptor alpha-1, secreted protein, acidic and rich in cysteine/osteonectin, and thrombospondin 4. We hypothesize that the abnormal Ca(2)+ influx in dystrophin- and alpha-sarcoglycan-deficient myofibers leads to altered developmental programming of developing and regenerating myofibers. The finding of upregulation of HLA-DR and factor XIIIa led to the novel identification of activated dendritic cell infiltration in dystrophic muscle; these cells mediate immune responses and likely induce microenvironmental changes in muscle. We also document a general metabolic crisis in dystrophic muscle, with large scale downregulation of nuclear-encoded mitochondrial gene expression. Finally, our expression profiling results show that primary genetic defects can be identified by a reduction in the corresponding RNA.
Figures






Comment in
-
Muscular dystrophy meets the gene chip: new insights into disease pathogenesis.J Cell Biol. 2000 Dec 11;151(6):F43-5. doi: 10.1083/jcb.151.6.f43. J Cell Biol. 2000. PMID: 11121429 Free PMC article. No abstract available.
References
-
- Barbiroli B., Funicello R., Iotti S., Montagna P., Ferlini A., Zaniol P. 31P-NMR spectroscopy of skeletal muscle in Becker dystrophy and DMD/BMD carriers. Altered rate of phosphate transport. J. Neurol. Sci. 1992;109:188–195. - PubMed
-
- Bonnemann C.G., Modi R., Noguchi S., Mizuno Y., Yoshida M., Gussoni E., McNally E.M., Duggan D.J., Angelini C., Hoffman E.P. Beta-sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat. Genet. 1995;11:266–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

