Interaction of human DNA topoisomerase I with G-quartet structures
- PMID: 11121473
- PMCID: PMC115246
- DOI: 10.1093/nar/28.24.4832
Interaction of human DNA topoisomerase I with G-quartet structures
Abstract
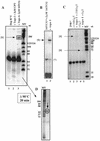
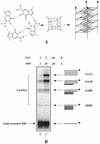
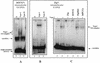
Because of their role in the control of the topological state of DNA, topoisomerases are ubiquitous and vital enzymes, which participate in nearly all events related to DNA metabolism including replication and transcription. We show here that human topoisomerase I (Topo I) plays an unexpected role of 'molecular matchmaker' for G-quartet formation. G-quadruplexes are multi-stranded structures held together by square planes of four guanines ('G-quartets') interacting by forming Hoogsteen hydrogen bonds. Topo I is able to promote the formation of four-stranded intermolecular DNA structures when added to single-stranded DNA containing a stretch of at least five guanines. We provide evidence that these complexes are parallel G-quartet structures, mediated by tetrads of hydrogen-bonded guanine. In addition, Topo I binds specifically to pre-formed parallel and anti-parallel G4-DNA.
Figures

References
-
- Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. - PubMed
-
- Nitiss J.L. (1998) Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta, 1400, 63–81. - PubMed
-
- Pommier Y., Pourquier,P., Fan,Y. and Strumberg,D. (1998) Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta, 1400, 83–105. - PubMed
-
- Redinbo M.R., Stewart,L., Kuhn,P., Champoux,J.J. and Hol,W.G. (1998) Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science, 279, 1504–1513. - PubMed
-
- Sen D. and Gilbert,W. (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its applications for meiosis. Nature, 334, 364–366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

