Characterization of the B lymphocyte-induced maturation protein-1 (Blimp-1) gene, mRNA isoforms and basal promoter
- PMID: 11121475
- PMCID: PMC115243
- DOI: 10.1093/nar/28.24.4846
Characterization of the B lymphocyte-induced maturation protein-1 (Blimp-1) gene, mRNA isoforms and basal promoter
Abstract
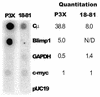
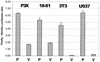
Blimp-1 is a transcriptional repressor that is both required and sufficient to trigger terminal differentiation of B lymphocytes and monocyte/macrophages. Here we report the organization of the mouse Blimp-1 gene, an analysis of Blimp-1 homologs in different species, the characterization of Blimp-1 mRNA isoforms and initial studies on the transcription of Blimp-1. The murine Blimp-1 gene covers approximately 23 kb and contains eight exons. There are Blimp-1 homologs in species evolutionarily distant from mouse (Caenorhabditis elegans and Drosophila melanogaster) but no homolog was found in the unicellular yeast Saccharomyces cerevisiae. The three major Blimp-1 mRNA isoforms result from the use of different polyadenylation sites and do not encode different proteins. Run-on transcription analyses were used to show that the developmentally regulated expression of Blimp-1 mRNA in B cells is determined by transcription initiation. Multiple Blimp-1 transcription initiates sites were mapped near an initiator element and a region conferring basal promoter activity has been identified.
Figures







References
-
- Krenacs L., Himmelmann,A.W., Quintanilla-Martinez,L., Fest,T., Riva,A., Wellmann,A., Bagdi,E., Kehrl,J.H., Jaffe,E.S. and Raffeld,M. (1998) Transcription factor B-cell-specific activator protein (BSAP) is differentially expressed in B cells and in subsets of B-cell lymphomas. Blood, 92, 1308–1316. - PubMed
-
- Oliver A.M., Martin,F. and Kearney,J.F. (1997) Mouse CD38 is down-regulated on germinal center B cells and mature plasma cells. J. Immunol., 158, 1108–1115. - PubMed
-
- Turner C.A. Jr, Mack,D.H. and Davis,M.M. (1994) Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell, 77, 297–306. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

